Of all the building blocks of life, glycans are the most abundant and diverse family of organic molecules, which informational content potentially exceed that of nucleic acids, proteins and lipids. Their complexity originates from the innumerable regio- and stereo-diverse ways in which simple sugar monomers can assemble to form highly branched polymers. This dense array of glycan covers the surface of every living cell, controlling many of their biological functions. Because of the wide diversity of structures that can be generated by assembling individual monosaccharides, they have been compared to the letters of an alphabet. As letters can combine in words, saccharides assemble on the surface of the cell generating a “biochemical language”, in which informational properties are encoded in the 3D structure of glycans themselves. The “biochemical language” has been described as following the so-called “glycocode”. A fundamental role in deciphering the “sugar code” is played by lectins, (Gabius et al., 2011) carbohydrate-binding proteins lacking enzymatic activity, which occur in all organisms. It’s not by chance that the name “lectin” derives from the Latin “legere”, to read/to select, referring to their ability to recognise glycans and to translate their structural information into function. Lectins have been intensively studied: they have been isolated and characterized mostly in plants, followed by animals, viruses, bacteria, fungi and yeasts. Prone to yield single crystals of good quality, there are many X-ray structures of lectin available (source:https://www.unilectin.eu/unilectin3D/). To have an update on state of the art, UniLectin3D is a free and useful database that classifies all known lectins in different families based on their origin, the structure of their carbohydrate-binding domains, or monosaccharide specificity. (Bonnardel et al., 2019)
Lectin-carbohydrate interactions have been studied in several biological processes of critical importance, such as immune recognition, cell recruitment, signal transduction, infection, and fertilization (Imberty & Varrot, 2008; Pang et al., 2011; Gabius, 2006; Meiers et al., 2019). Lectins are often involved in host-pathogen recognition and tissue adhesion. Consequently, they have become appealing targets for the design of glyco-drugs that aim at interfering with such processes (Audfray et al., 2013). Especially for the role they play in disease, understanding lectin specificity and function has become a priority in carbohydrate research.
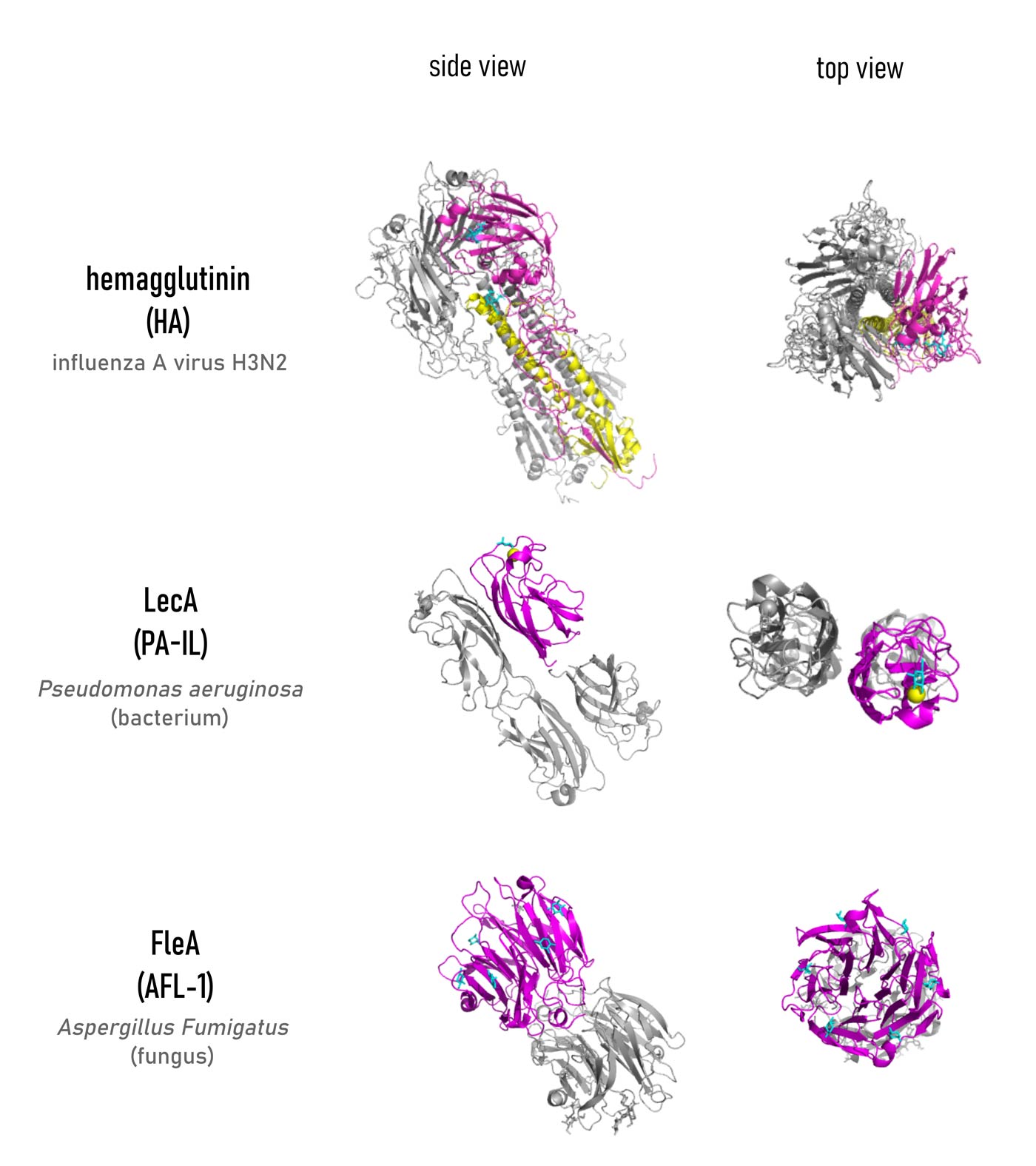
Since the first lectin was isolated in 1972, (Hardman & Ainsworth, 1972) about 300 others, have been revealed and appeared in different quaternary structures, although sharing a relatively limited number of folds and binding site topologies. An in-depth analysis of the binding mode of carbohydrate ligands in the lectin binding site is essential to understand the mechanism of recognition. Especially crystallographic and isothermal titration calorimetry (ITC) studies have been conducted to gather information about the structure of lectin-ligand complexes and the strength of their interaction. Possessing numerous hydroxyls, sugar epitopes can easily establish a series of hydrogen bonds (either direct or water-mediated) with the amino acids in the binding site. Such hydrogen bond network appeared to give the highest contribution to the enthalpy (ΔH) of binding. Additionally, metal coordination (such as Ca2+ chelation for C-type lectins), hydrophobic and ionic (for sialic acid-binding lectins) interactions, contribute to constructing a specific arrangement of contacts that binds the glycan with high specificity (Varrot & Blanchard, 2011). This specificity is what allows lectins to read the “glycocode” (Gabius et al., 2011).
Despite the perfect fit of the ligand inside the binding pocket, the affinity of monosaccharides for such proteins is quite low, frequently in the millimolar range (Imberty et al., 2005). Some oligosaccharides have binding affinities in the micromolar range, due to their ability to establish a higher number of hydrogen bonds. Such a feature results in a more favourable ΔH upon binding (although still paying an increased entropy penalty because of decreasing flexibility). To compensate for such a flaw, nature organized a scenario in which simultaneous low-affinity interactions between carbohydrate ligands and their receptors are possible, leading to stronger affinity. Scientists agree in describing this effect as “multivalency” and have underlined its importance in many publications (Pieters, 2009; Roy et al., 2016; Dam & Brewer, 2008; Bernardi et al., 2013; Wolfenden & Cloninger; 2011; Compain, 2020).
To learn more about glycans, watch “Chemical Glycobiology” – Carolyn Bertozzi (UC Berkeley)
