UPDATE July 2016
An updated version of the molecular visualization program UnityMol/SweetUnityMol is available. It is fully compatible with the Symbol Nomenclature for Glycan (SNFG) colour coding published in Glycobiology, 2015, 25 (12) 1323-1324.
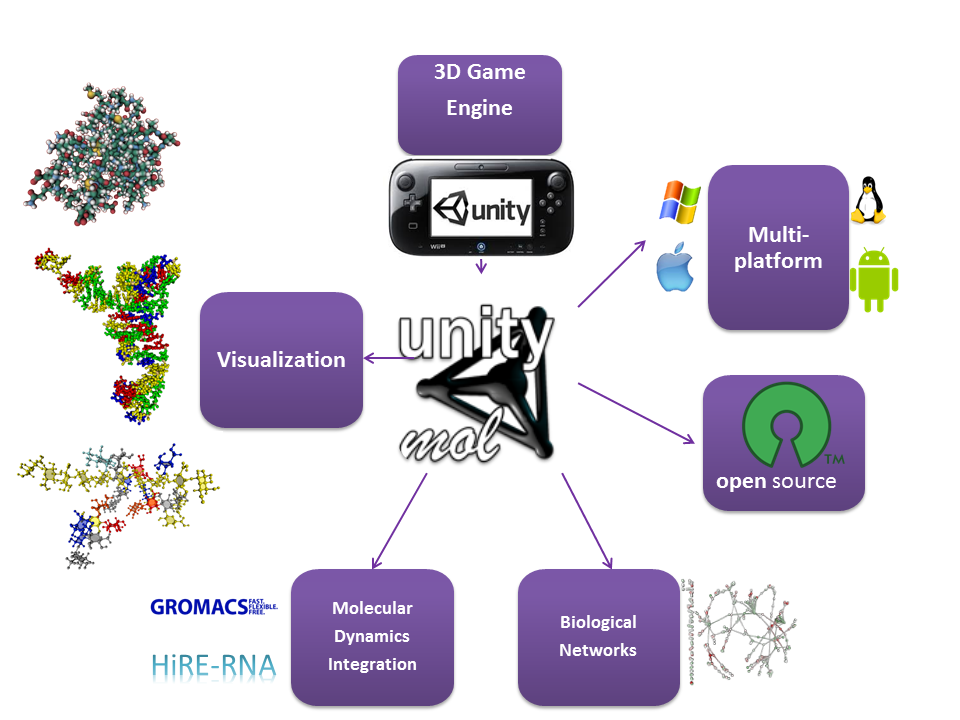
The software has been initially developed based on revision 676 of UnityMol and version4.5.2f1 of Unity3D. All source code was implemented using C# and Cg languages built into Unity 3D and is available along with executables for Mac, Windows and Linux platforms on the sourceforge project website. Current UnityMol versions run within version 5.3.2f1 of Unity3D.
The software is freely available on the sourceforge project
The SweetUnityMol User Manual provides documentation, input files, and series of illustrations. It can be Downloaded
PRESENTATION December 2014
A molecular visualization program tailored to deal with the range of three-dimensional structures of complex carbohydrates and polysaccharides, either alone or in their interactions with other bio-macromolecules, has been developed using advanced technologies elaborated by the video games industry.
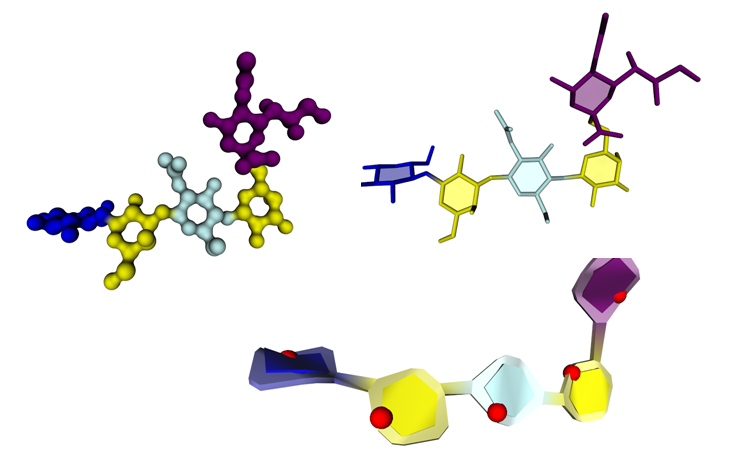
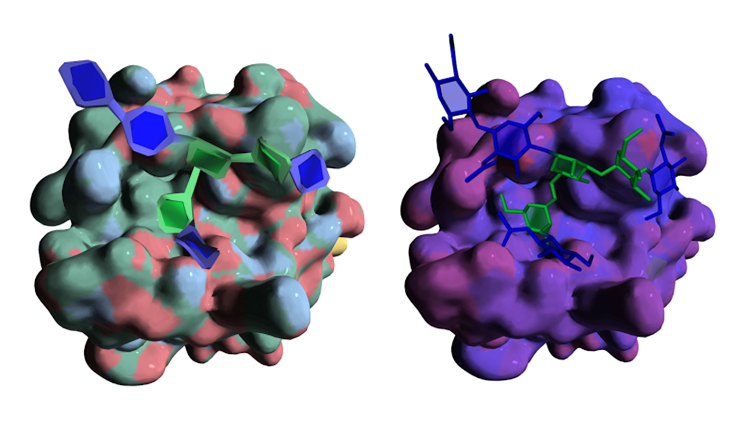
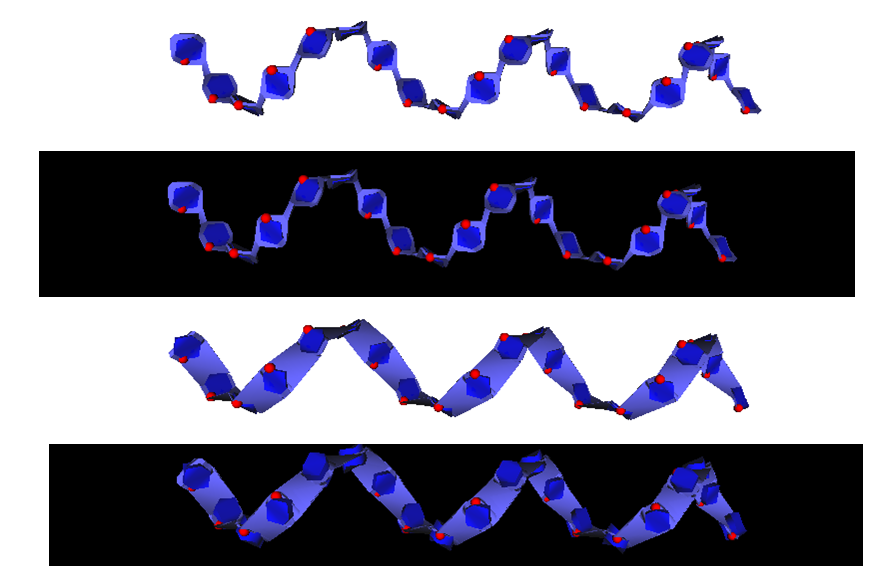
All the specific structural features displayed by the simplest to the most complex carbohydrate containing molecules have been taken into account and can be conveniently depicted. This concerns the monosaccharide identification and classification, conformations, location in single chain or multiple branched chains, depiction of secondary structural elements, and the essential constituting elements in very complex structures. In all these instances, particular attention was given to cope with the accepted nomenclature and pictorial representation used in carbohydrate chemistry, biochemistry and glycobiology. The present achievement provides a continuum between the most popular ways to depict the primary structures of complex carbohydrates to visualizing their 3D structures while giving the users many options to select the most appropriate modes of representations including new features such as those provided by the use of textures to depict some molecular properties in an intuitive way.
The present approach emphasizes how advanced technologies developed by the video game industry can be used and extended to the delivery of easily modifiable and extensible scientific research tools. These developments are incorporated in a stand-alone viewer capable of displaying molecular structures, bio-macromolecule surfaces, and complex interactions of bio-macromolecules, with powerful, artistic and illustrative rendering methods. They result in an open source software compatible with multiple platforms i.e. Windows, MacOS and Linux operating systems, web pages and producing publication-quality figures.
To demonstrate their utility, the algorithms and visualization enhancements are exemplified using a variety of carbohydrate molecules, from glycan determinants to glycoproteins and complex protein-carbohydrate interactions, as well as very complex mega-oligosaccharides and bacterial polysaccharides and multi-stranded polysaccharide architectures.
All source code was implemented using C# and Cg languages built into Unity 3D and executables for Mac, Windows and Linux platforms are provided.
The authors

Serge Perez

Thibault Tubiana

Anne Imberty

Mark Baaden
