The phenomenon of surface plasmon resonance (SPR) was observed for the first time by Wood more than a century ago (Wood, 1902). Since then, the number of publications on the topic increased exponentially (Rich et al., 2011), until reaching several thousand today. Only 90 years after the discovery of the theory of SPR, the first BiacoreTM instrument was commercialized by a Swedish company now part of the GE Healthcare group. More and more advanced instruments have nowadays become available from various companies around the world.
The SPR technology allows us to monitor receptor-ligand interactions in real-time. Similarly to ELLA, one biomolecular partner is immobilized on a sensor chip surface, but there is no need for enzyme labelling in SPR detection. Conversely, the signal is generated by the change in the refractive index close to the sensor surface, as the ligand binds. The chip consists of a glass layer, a thin gold film, and a polymeric surface (generally dextran) with a bio-specific coating. The device is composed not only of the sensor chip, but also an optical device and a microfluidic cartridge. When a light beam hits the surface of the chip, it is reflected onto the detector device. At a certain resonance angle, the energy of the beam is transferred to the surface electrons of the metal layer, called plasmons, making them resonate. Integrated into the SPR machinery, a continuous flow of analyte in solution is injected on the surface, allowing the biomolecular pair to get in contact. Surface plasmons are sensitive to every slight change in their environment, which results in a variation of the refractive index of the media in which light is travelling. Such change influences the angle of reflectance too, so that the intensity of reflected light decreases at a fix detector position. The more material gets bound to the surface, the more significant variation of refractive index is observed. (Walker et al., 2003)
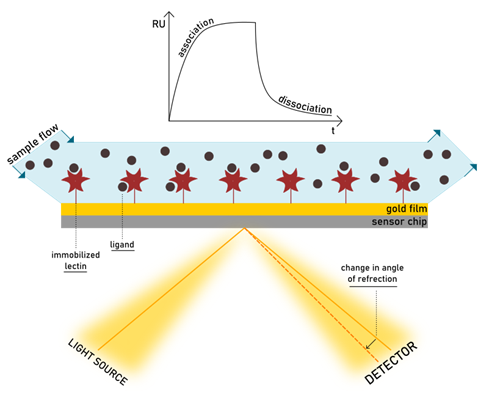
In an SPR experiment, the reflected light is monitored continuously, and the interaction is detected as it forms. Any adhesive event can be followed in the SPR sensorgram, expressed in resonance units (RU) per time. The increase of mass on the surface during ligand-protein association causes the SPR response to increase, while dissociation is detected as a decrease in response. A visual representation of the theory of SPR can be found at the links https://youtu.be/sM-VI3alvAI and https://youtu.be/o8d46ueAwXI, as explained for the BiacoreTM or the Biosensing Instruments products.
Since SPR allows to study a biomolecular interaction over time, it readily offers insights on its kinetic. The value of kinetic constants (kon and koff) are readily available from the sensorgram. The signal at equilibrium gives access to the thermodynamic parameter Kd. The direct access to the kinetic profile, and to some extent to the thermodynamic information, is unique for SPR in the same and opposite way that it is for ITC. Generating both equilibrium and kinetic data, SPR is powerful in determining fundamental factors for drug optimization and the study of biological events. Notably, the easy achievement of kinetic parameters is important. If the Kd is a well-established representative of the strength of binding and extensively exploited in drug design, the elaboration of kinetic information is less straightforward but necessary. The residence time, dictated by koff, has been assigned a key role in influencing biological activity (Bernetti et al., 2019).
The great informational potential of SPR is counterbalanced by some troublesome elements to consider. A frequent obstacle in the kinetic analysis is represented by the transport mass limitation. In inconvenient situations, the diffusion of analyte from the solution to the surface of the chip becomes rate-limiting for the process in observation. In such conditions the apparent on- and off- rates do not correspond to the intrinsic ones. This observation happens when the flux of analyte is slow, the density of the immobilized biomolecule is high, or when the interaction has a fast association (high kon). These aspects should be controlled in the experiment, when possible. For example, the right flow conditions can be verified by analysing the effect of different sample flow rates on the experimental binding rates, which should not be affected by the changes.
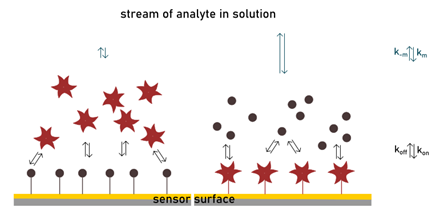
Controlling the density of the matrix is indeed a more delicate task. If on the one hand, high surface capacities risk falling into mass transport problems, diversely low densities can incur in sensitivity issues and non-specific binding effects. The need to find a good compromise between signal strength and binding capacity is a disadvantage of SPR. The selection of a suitable matrix is critical. The choice is vast: nowadays many surfaces are available, with hydrophilic or hydrophobic properties, trying to mimic a biological context (e.g. a cellular membrane). Even more importantly, it is possible to optimize the surface, adapting it to one’s purpose. Surface loading can be controlled using different types of chemistry. Immobilization of a biomolecule on the matrix may require complex or standard procedures and can exploit covalent or non-covalent interactions. Different assay setting can be implemented to study lectin-ligand binding. One consists of immobilizing the lectin on the surface. This is typically done via a standard amine coupling. The delicate aspect of this procedure is the difficulty in controlling the orientation and assure that the binding site is maintained intact. Using the streptavidin-biotin system for protein ligation can usually solve this problem. A direct binding assay provides information on specificity, thermodynamic and kinetic features of the biological event. Such assay can be performed when a flow of glycans is injected on the matrix which exposes the lectin, but also on the reverse way.
In general, low molecular weight ligands provide low mass variation upon binding, therefore low sensitivity. For this reason, glycoside ligands are typically immobilized when aiming at high signal-to-noise ratios. In this second assay setting, several techniques are also available for the immobilization of carbohydrates or glycoproteins on the chip. Again, functionalization should not modify pharmacophore moieties, otherwise interfering with the binding interaction. Carbohydrate-immobilized assays can also be employed in glycoprofiling (i.e. studying the oligosaccharide sequence present, for example, on a glycoprotein), when lectin binding is analysed after treatment of the surface with glycosidases (Walker et al., 2003).
More frequently, the immobilization of glycosides on the surface is used in competitive binding assays. In inhibition studies, a known ligand is immobilized on the surface, and a more valuable synthetic glycoside is added with the lectin in the buffer. Equilibrium responses are used to determine IC50s. Importantly, the same chip can be reused in more experiments, allowing to screen drug candidates while minimizing variability between each independent investigation. The evaluation of multivalent glycosides is, of course, possible in these conditions. However, the accurate determination of kinetic parameters is complicated (Schlick & Cloninger, 2010). Given the heterogeneous nature of binding events with the surface and the compound in solution, it is difficult to deconvolute a single binding mechanism from the whole process.
In Nature, glycans exist as a dense decoration on the surface of the cell. The interaction with their respective receptor occurs at the interface with the extracellular matrix. The competitive situation well simulates many carbohydrate-lectin interactions, since the chip surface mimics the glycocalyx and the binding with an inhibitor happens in solution. Measuring interactions on a solid surface like in SPR can also clarify the nature of multivalency effects. Avidity enhancements measured by SPR have shown to be dependent on which partner is immobilized within the biomolecular pair as well as the density of immobilization and the oligomeric state of the protein. When the glycoside ligand is part of the matrix, lectin binding is stronger compared to when the set-up is reversed (Shinohara et al., 1997). The significant increase in affinity constant is mainly related to changes in the dissociation rate. The value of koff is much lower in the carbohydrate-immobilized assay and enhanced by lectin oligomerization. This, together with fast kon typical for lectin-carbohydrate interactions, contributes to creating a non-homogeneous state that is limited by transport mass issues mentioned earlier (De Mol & Fischer, 2010). In such conditions, the rebinding effect becomes extremely large and cannot be neglected. Multivalent ligands (or multiple ligands on a surface) positively utilize this mechanism to modulate apparent affinity: a lectin will be easily entrapped in the matrix and unlikely it will leave it.
Since many factors can affect binding parameters, the SPR experiment needs to be adequately arranged. This means optimizing the density of immobilized material, concentrations of a partner in solution, injection times and buffers. Another drawback is that the sensor chips are expensive and have a limited possibility of reuse.
Even though SPR has critical points, it is characterized by many advantageous features too. Remarkably, SPR is one of the most sensitive label-free techniques available for analysing lectin-sugar interactions with affinities up to picomolar range. The amounts of compounds needed are low: working volumes are generally in the range of 50–150 μL. Thanks to the miniaturization offered by the manufactured automated instruments, many binding assays can be simultaneously evaluated in real-time. New frontiers in SPR analysis are aiming at the elaboration of coupled analytical techniques to achieve more detailed information over complex mixtures quickly. Thanks to avant-garde technologies, concomitant quantitative and qualitative analysis are now possible (Safina, 2012).
For example, SPR has been integrated with mass spectrometry to obtain valuable structural information from molecular weight and fragmentation patterns. SPR imaging is another variation of the classic SPR machine which incorporates microscopy (a more detailed explanation of SPRi can be found at https://youtu.be/c4SrPzfOZNE). This extension permits to observe an array of bio-interactions simultaneously, adding a visual dimension to the measurement. However, the great convenience of using more advanced instrumentation goes along with its higher costs.
