Two recently published monographs, “A road-map for Glycoscience in Europe” and “Transforming Glycoscience : A Roadmap for the future” published respectively under the auspices of the European Science Foundation and the National Academies USA, identified a selected number of goals. One of particular importance was the need for the “establishment of long-term databases and bioinformatics and computational tools to enable accurate carbohydrate and glycoconjugate structural predictions”. One This recommendation highlights a major challenges facing Glycoscience is namely, the development and implementation of robust and validated informatics toolbox enabling accurate and fast determination of complex carbohydrate sequences extendable to 3D prediction, computational modeling, data mining, and profiling.
Another key challenge for Glycoscience is to pull it out of its isolation. Glyco-molecules are functionally important in many biological processes yet their occurrence, let alone their role, is rarely considered. Nonetheless the recommended shift towards bioinformatics as stated above, is a chance to bridge Glycoscience with other fields. Indeed, the concomitant expansion of stable and integrated databases, cross-referenced with popular bioinformatics resources should contribute to connecting glycomics with other –omics.
Proteomics, transcriptomics and genomics are at the core of biology and medicine whereas lipidomics, metabolomics and glycomics seldom are taken into account.
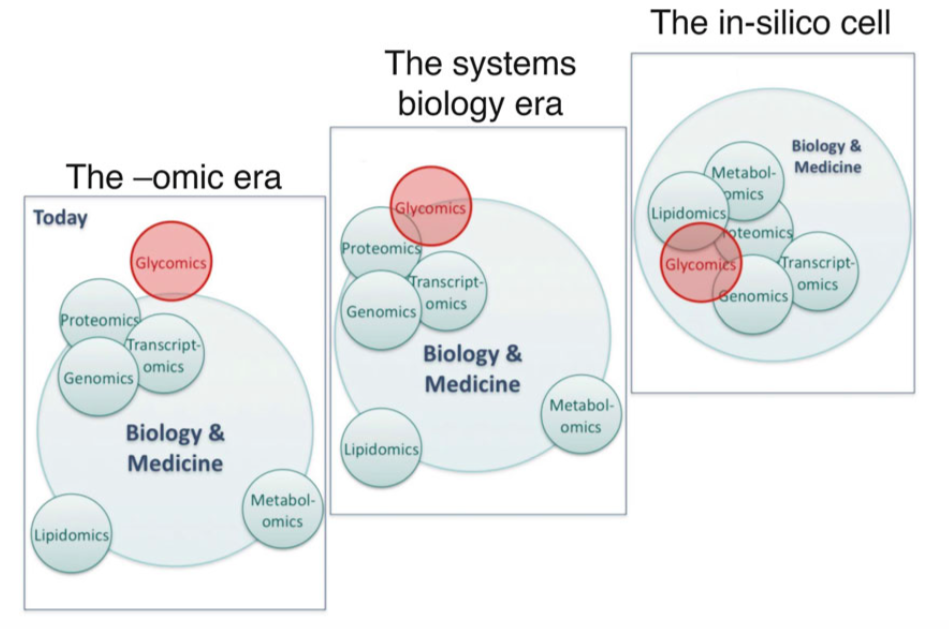
Bioinformatics played an essential role in the integration of genomics, proteomics and transcriptomics (Manzoni et al., 2018). The development of an integrated approach to assemble and annotate newly sequenced genomes from transcriptome and proteome data is just one example of the importance of bioinformatics (Prasad et al., 2017). However, other issues like the extent of proteoforms (Aebersold et al., 2018) or that of the metabolome still hamper data integration, leaving the puzzle incomplete. Furthermore, several other -omics fields such as lipidomics or glycomics are remote in this picture mainly because of technical and knowledge gaps. This is particularly the case in glycomics that remains internally fragmented and misses solid bioinformatics support. Only the integration of all the ’omics’ will lead to a in-silico simulation on a living cell.
The upcoming scientific production in Glycoscience amounts to the yearly publication of about 70,000 research articles. Amplified by the availability of sophisticated and powerful high-performance computing and searching capacities, the space of accessible information has substantially increased such that data mining opens new prospects of discovery. This new view not only emphasises the worth of analyzing raw data from published work, but also points at untapped wealth that may be harvested in collected data sets from which extracted information can be transformed into knowledge. The field of structural glycobiology has partially benefited from such advances with the development of tools and databases for structural analysis of carbohydrates. A variety of other online resources mainly in the form of databases covering glycan and glycoproteins structures, enzymes responsible for their biosynthesis and degradation, glycan binding to human pathogens, glyco-epitope and their antibodies,… has been developed by independent research groups, worldwide.
The present chapter covers the description of resources, in the form of tools and databases, that are freely available (most of them being web-based) and are regularly updated and improved. With the aim of facilitating glycoscience research, we have clustered these different tools according to their major field of applications. As a result, the following entries can be accessed : Portals ; Genome and Glycome ; Representations ; Experimental Results ; Glycan ; Glycoproteomics ; Functional Glycomics ; Glycolipids ; CAZYmes & Polysaccharides.
