With the aim of facilitating glycoscience research, we have identified the tools and databases that are freely available on the internet and are regularly updated and improved. They have been clustered according to the major fields of applications: : Portals ; Genome and Glycome ; Representations ; Experimental Results ; Glycan ; Glycoproteomics ; Functional Glycomics ; Glycolipids ; CAZYmes & Polysaccharides.
PORTALS
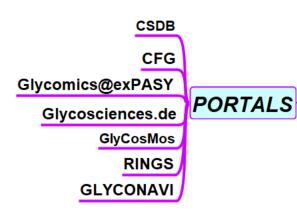
- Carbohydrate Structure Data Base http://csdb.glycoscience.ru/database/core/help.php?topic=rules CSDB covers information on structures and taxonomy of natural carbohydrates published in the literature and mostly resolved by nuclear magnetic resonance (NMR). CSDB is composed of two parts: Bacterial & Archeal (BCSDB) and Plant & Fungal (PFCSDB).
- Consortium for Functional Glycomics- CFG http://www.functionalglycomics.org/ The CFG serves to combine the expertise and glycomics resources to reveal functions of glycans and glycan-binding proteins (GBPs) that impact human health and disease. The CFG offers resources to the community, including glycan array screening services, a reagent bank, and access to a large glycomics database and data analysis tools.
- ExPASy https://www.expasy.org/glycomics Glycomics@ExPASy is the glycomics tab of ExPASy, the server of SIB Swiss Institute of Bioinformatics. It centralizes web-based glycoinformatics resources developed within an international network of glycoscientists.
- GlyCosMos Portal https://glycosmos.org GlyCosmos Portal Development entails the integration of more diverse –omics data including glycogenes, glycoconjugates such as glycoproteins and glycolipids, molecular structures, and pathways.
- GLYCONAVI http://www glyconavi.org/ GlycoNAVI is a web site for carbohydrate research. It consists of the ‘GlycoNAVI DataBase’ for molecular information of carbohydrates, and chemical reactions of carbohydrate synthesis, the ‘Route Searching System for Glycan Synthesis’, and ‘GlycoNAVI Tools’ for editing two-dimensional molecular structure of carbohydrates.
- Glycoscience.de http://www.glycosciences.de The Glycosciences.de web portal provides databases and tools to support glycobiology and glycomics research. Its main focus is on 3D structures, including 3D structure models as well as references to PDB entries that feature carbohydrates
- RINGS http://rings.t.soka.ac.jp/ RINGS is a web resource providing algorithmic and data mining tools to aid glycobiology research
GENOME – GLYCOME
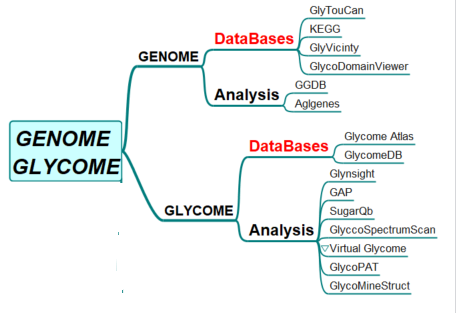
- GlycoDomainViewer https://glycodomain.glycomics.ku.dk/
GlycoDomainViewer: a bioinformatics tool for contextual exploration of glycoproteomes. It presents a first map of the human O-glycoproteome with almost 3000 glycosites - Glycome Analytics Platform GAP https://bitbucket.org/scientificomputing/glycome-analytics-platform The Glycome Analyti
- GlycomeAtlasV5: Visualization of glycome profiling data on human and mouse tissue samples http://rings.t.soka.ac.jp/GlycomeAtlasV5/index.html
- GlycoGeneDataBase GGDB https://acgg.asia/ggdb2 Glycogene includes genes associated with glycan synthesis such as glycosyltransferase, sugar nucleotide synthases, sugar-nucleotide transporters, sulfotransferases, etc.
- GlycoMinestruct: http://glycomine.erc.monash.edu/Lab/GlycoMine_Struct/ a new bioinformatics tool for highly accurate mapping of the human N-linked and O-linked glycoproteomes by incorporating structural features
- Glynsight https://glycoproteome.expasy.org/glynsight/ Glynsight offers visualization and interactive comparison of glycan expression profiles. The tool was initially developed with a focus on IgG N-glycan profiles but it was extended to usage with any experiment, which produces N- or O-linked glycan expression data.
- GlycoSpectrumScan https://github.com/wliu1197/glycospectrumscan A web-based bioinformatic tool designed to link glycomics and proteomics analyses for the characterization of glycopeptides. GlycoSpectrumScan is MS-platform independent, freely accessible, and profiles glycopeptide MS data using beforehand separately acquired released glycan and proteomics information. Both N- and O-glycosylated peptides as well as multiply glycosylated peptides can be analyzed
- Glytoucan https://glytoucan.org/ GlyTouCan is the international glycan structure repository.
- GlyVicinity http://www.glycosciences.de/tools/glyvicinity/ Analysis of Amino Acids in the Vicinity of Carbohydrate Residues contained in the Protein Data Bank (PDB)
- KEGG GLyCan database https://www.genome.jp/kegg/glycan/
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a bioinformatics resource for linking genomes to life and the environment. - SugarQb http://www.imba.oeaw.ac.at/SugarQb SugarQb enables genome-wide insights into protein glycosylation and glycan modifications in complex biological systems. This is a collection of software tools (Nodes) which enable the automated identification of intact glycopeptides from HCD‐MS/MS data sets, using commonly use peptide-centric MS/MS search engines
- Virtual Glycome https://virtualglycome.org/ This website is focused on presenting selected computational tools and experimental resources that can be used to better understand the processes regulating cellular glycosylation at multiple levels.
