The collection of tools and databases described in detail in the previous sections can be grouped into two categories. They can either be dedicated to solving a specific question or be used in an integrative way in several applications. In the context of exploring and understanding the biological functions where glycans are involved, toolboxes are required to navigate, investigate and correlate data. One such a facility is offered by the crosslinks of databases such as GlyS3, SugarBindDB, GlyConnect and the respective crosslinks to UniProt. A user can seek to establish the consistency of interactions taking place at the cell surface. In the following, three possible use cases are brought to the practitioner.
From MS to glycoprotein features.
This toolset is designed to match the expected boost of glycoproteomics (glycan composition at specific sites on complex mixtures of glycoproteins) data that is currently just reaching high throughput level. An example of how to integrate some of these dedicated tools for extracting glycoprotein features from MS data is shown below
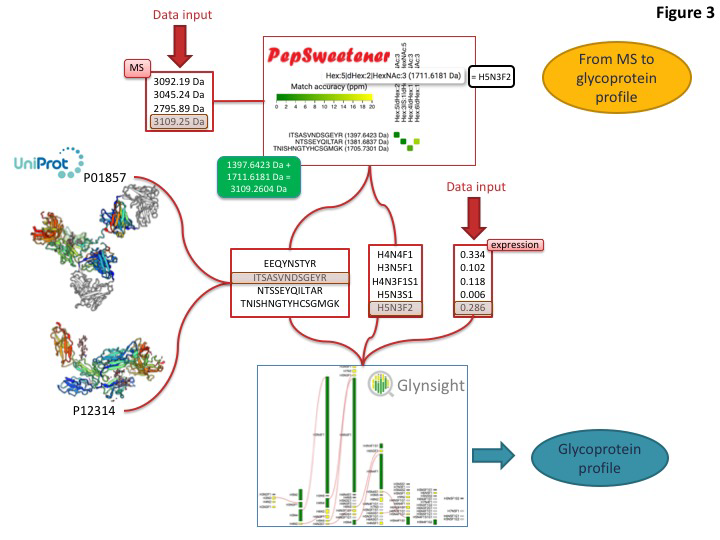
Predominant precursor masses in the MS spectra can be input into PepSweetener. This software supports the manual annotation of intact glycopeptides, using custom web visualization regardless of the instrument that produced the data. An interactive heat-map chart displays the results; it features the combined mass contributions of theoretical (usually tryptic) peptides and attached glycan compositions. The variations in tile colours correspond to ppm deviations from the query precursor mass. Annotation can be refined through glycan composition filtering, sorting by mass and tolerance, and checking MS-MS data consistency via an in silico peptide fragmentation diagram (in-house fragmentation tool common with that of UniCarb-DB). PepSweetener is mainly designed as a complement or extension to software being developed for automatic analysis of glycoproteomics MS data and avoiding their dependency on a set workflow or type of instrument. The outcome of this study will guide the presentation of the Glycomics@ExPASy toolbox towards a more informative and instructive section on MS-based glycoproteomics data analysis tools.
Exploring glycoprotein features.
Current global glycome profiling experiments generate one or more set(s) of glycan compositions and structures with their respective expression on a protein, in a tissue or a cell. Tools and databases in Glycomics@ExPASy can be combined to explore distinctive glycan features that characterise glycoproteins as shown in figure 3. In this case, the entry point of the workflow is GlyConnect to which a list of glycan compositions is submitted. The GlyConnect search tool will retrieve the possible related glycan structures and the proteins that have been reported to have these compositions/structures attached and stored in the databases. A conceptual map displays the results; the compositions sit in the middle and connect glycan structures and associated glycoproteins, respectively on the right and the left sides on the Figure below.
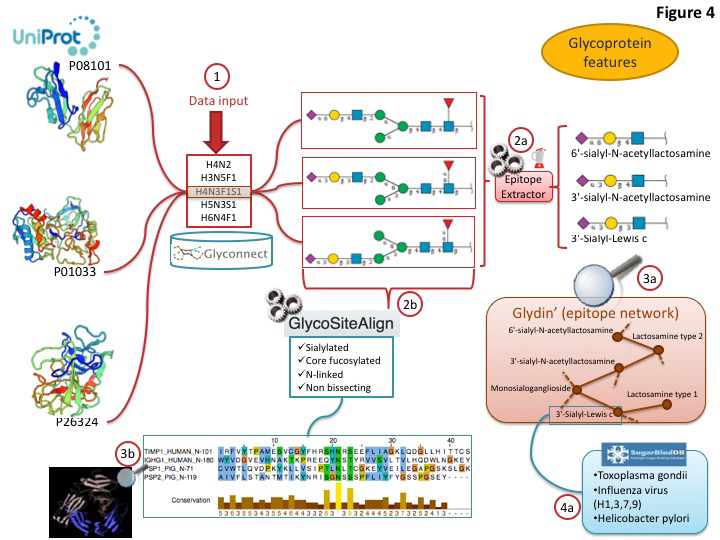
This visualization is well suited for understanding the potential relations between proteins and glycans. Activating the integrated EpitopeXtractor function provides a selection of glycan structures.
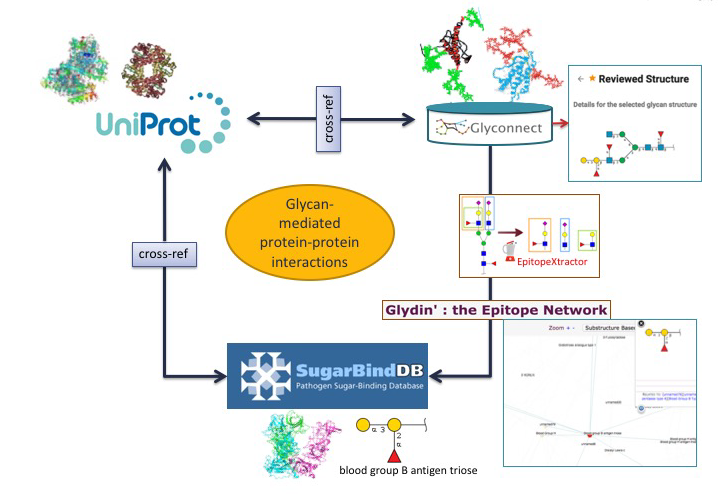
Glycan-mediated protein-protein interactions.
Using another combination of tools and databases in Glycomics@ExPASy, potential correlations between a glycan-binding protein (GBP) of a pathogen a host glycoprotein and a glycan structure can be made.
In the first scenario, the starting point is a glycoepitope recognised by a specific GBP, a bacterial lectin described in SugarBindDB.
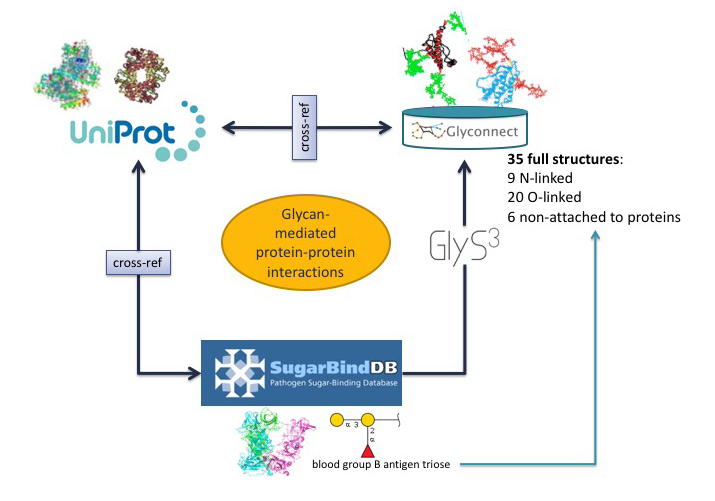
The blood group B antigen triose illustrates this point. A binding event in this database is always formed by A pair composed of a GBP/lectin and a glycoepitope part of a glycan present on the host surface defines a binding event in the database. Whenever possible, further information of a GBP/lectin is available via cross-reference to UniProt. The glycoepitope can be used as an input of the GlyS3 substructure search tool to match the full structures stored in GlyConnect that contain this specific ligand. The list of glycan structures retrieved by GlyS3 can be explored in GlyConnect that reports relationships between glycans and glycoproteins.
The second scenario starts from a glycan structure in GlyConnect and relies on its reported relationships with glycoproteins.

The figure shows an example of a reviewed N-linked glycan structure. GlyConnect also offers the option of running EpitopeXtractor to generate a selection of glycoepitopes contained in this starting glycan. Leveraging the binding data in SugarBindDB, the obtained glycoepitopes can be associated with a collection of GBPs/lectins that recognize one or more of these glycoepitopes. In the end, the workflow allows the selection of GBPs that could possibly interact with the glycoproteins on which the starting glycan has been reported to be attached. Cross-references of both glycoproteins in GlyConnect and GBPs/lectins in SugarBindDB to UniProt can be used to further rationalise potential interacting partners.