TOTO_August_2024
Structural Glycobioinformatics
GLYCOTwinning Scientific Network Meeting & 2nd Summer School 2024
Polys Glycan Builder: An Online Application for Intuitive Construction of 3D Structures of Complex Carbohydrates
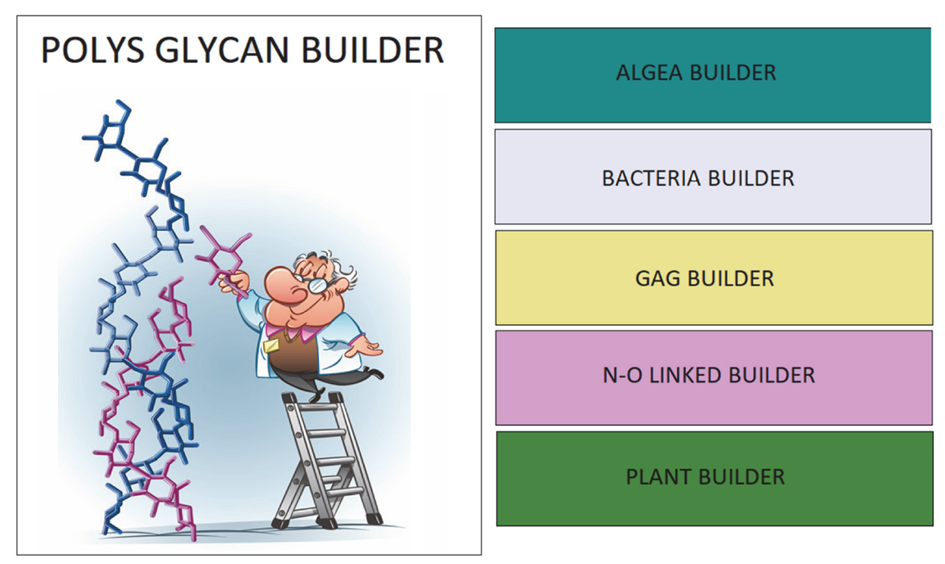
Serge Perez University of Grenoble Alpes, CNRS, Centre de Recherches sur les Macromolecules Vegetales, Grenoble, France spsergeperez@gmail.com; glycopedia.eu Abstract Polys-Glycan-Builder is a web-based, user-friendly interface for building 3D structures of complex glycans and polysaccharides. The construction follows an intuitive scheme as close as possible to how glycoscientists draw their structures’ sequences. The software translates a […]
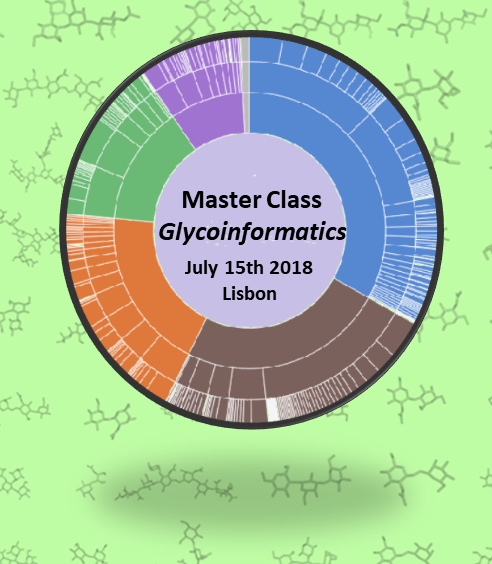
Masterclass Glycoinformatics
Information searching and data extraction are benefitting from the formidable contribution of computational science and technology. To manage data, and to make information accessible to a wider scientific community, publicly available databases and modeling tools for glycoscience have been developed, both in the field of glycochemistry and glycobiology. These tools are reaching maturity and standards […]
Program + Handouts

The proposed “Master Class in Glycoinformatics” aims at presenting and popularizing existing tools to a large community through practical courses given by leading scientists in the field. Information searching and data extraction are benefitting from the formidable contribution of computational science and technology. To manage data, and to make information accessible to a wider scientific […]
Presentation
UPDATE July 2016An updated version of the molecular visualization program UnityMol/SweetUnityMol is available. It is fully compatible with the Symbol Nomenclature for Glycan (SNFG) colour coding published in Glycobiology, 2015, 25 (12) 1323-1324. The software has been initially developed based on revision 676 of UnityMol and version4.5.2f1 of Unity3D. All source code was implemented using C# and Cg languages built into Unity […]
Round Table
Click on the vignette round_table.pdf
Databases
CAZY – Carbohydrate Active Enzymes – describes the families of structurally-related catalytic and carbohydrate binding modules (or functional domains) of enzymes that degrade, modify or create glycosidic bonds http://www.cazy.org/ Glycan Library – a list of approximately 830 lipid-linked sequence-defined glycan probes derived from diverse natural sources or chemically synthesised https://glycosciences.med.ic.ac.uk/glycanLibraryIndex.html GlycoBase 3.2 – a database […]
Tools
CASPER – a tool for calculating NMR chemical shifts of oligo- and polysaccharides http://www.casper.organ.su.se/casper/ Glycan Builder – a software library and set of tools to allow for the rapid drawing of glycan structures with support for all of the most common symbolic notation formats http://www.unicarbkb.org/builder GlycoDomainViewer – an online resource to study site glycosylation with […]
The Picture Dictionnary
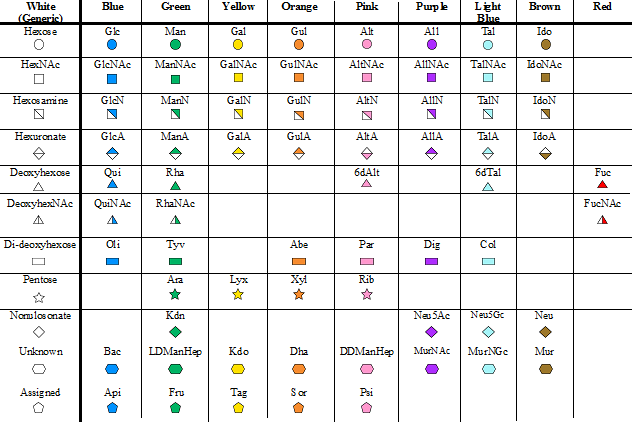
Content The Picture Dictionnary of Monosaccharide Symbols offers the pictorial representation of an extended series of monosaccharides. Each symbol follows the accepted rules in terms of shape and colors. Images of the symbols at the png format can be downloaded. Prepared lmages (png format) of glycosidic linkages is also provided.to be included in the representation […]
Presentation
This chapter offers a library of 128 monosaccharides which are presented throughout several levels of structural depiction, i.e. one dimension, two-dimension and three-dimension. The following three different families of monosaccharides are considered : hexoses, pentoses, ketoses. For a given configuration (D) or (L) monosaccharide can occur as α-pyranose, β-pyranose, α-furanose, β-furanose, i.e under eight different 3-Dimensional […]
Hexoses

GALACTOSE gets its name from Ancient Greek : “galakos” : milk αD-Galactofuranose αD-Galactofuranose β-D-Galactofuranose αD-Galactofuranose α-D-Galactopyranose α-D-Galactopyranose α-D-Galactopyranose α-D-Galactopyranose α-L-Galactofuranose β-L-Galactofuranose α-L-Galactofuranose α-L-Galactofuranose β-D-Galactofuranose β-D-Galactofuranose β-D-Galactofuranose β-D-Galactofuranose β-D-Galactopyranose β-D-Galactopyranose β-D-Galactopyranose β-D-Galactopyranose β-L-Galactofuranose β-L-Galactofuranose β-L-Galactofuranose β-D-Galactopyranose β-L-Galactopyranose β-L-Galactopyranose β-L-Galactopyranose β-D-Galactopyranose GLUCOSE gets its name from Ancient Greek : “gleukos” : sweet wine, must α-D-Glucofuranose α-D-Glucofuranose α-D-Glucofuranose […]
Pentoses

ARABINOSE gets its name from “Gum Arabic“, from which it was first isolated α-D-Arabinofuranose α-D-Arabinofuranose α-D-Arabinofuranose α-D-Arabinofuranose α-D-Arabinopyranose α-D-Arabinopyranose α-D-Arabinopyranose α-D-Arabinopyranose α-L-Arabinofuranose α-L-Arabinofuranose α-L-Arabinofuranose α-L-Arabinofuranose α-L-Arabinopyranose α-L-Arabinopyranose α-L-Arabinopyranose α-L-Arabinopyranose β-D-Arabinofuranose β-D-Arabinofuranose β-D-Arabinofuranose αD-Galactofuranose β-D-Arabinopyranose β-D-Arabinopyranose β-D-Arabinopyranose β-D-Arabinopyranose β-L-Arabinofuranose β-L-Arabinofuranose β-L-Arabinofuranose β-L-Arabinofuranose β-L-Arabinopyranose β-L-Arabinopyranose β-L-Arabinopyranose β-L-Arabinopyranose LYXOSE gets it name as an anagram of “Xylose“ α-D-Lyxofuranose […]
Ketoses

FRUCTOSE gets its name from Latin : “fructus“ α-D-Fructofuranose α-D-Fructofuranose α-D-Fructofuranose α-D-Fructofuranose α-D-Fructopyranose α-D-Fructopyranose α-D-Fructopyranose α-D-Fructopyranose α-L-Fructofuranose α-L-Fructofuranose α-L-Fructofuranose α-L-Fructofuranose α-L-Fructopyranose α-L-Fructopyranose α-L-Fructopyranose α-L-Fructopyranose β-D-Fructofuranose β-D-Fructofuranose β-D-Fructofuranose β-D-Fructofuranose β-D-Fructopyranose β-D-Fructopyranose β-D-Fructopyranose β-D-Fructopyranose β-L-Fructofuranose β-L-Fructofuranose β-L-Fructofuranose β-L-Fructofuranose β-L-Fructopyranose β-L-Fructopyranose β-L-Fructopyranose β-L-Fructopyranose PSICOSE gets its name from an alteration of pseudo (psi) Fructose“ α-D-Psicofuranose α-D-Psicofuranose α-D-Psicofuranose α-D-Psicofuranose α-D-Psicopyranose […]
Conformational Wheel 5 Membered Ring
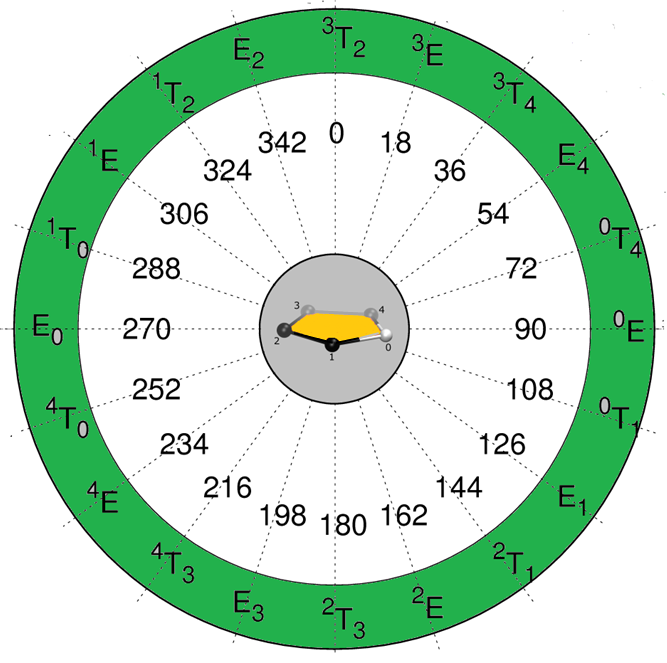
Five Membered-ring 3D Structures around the Conformational Wheel The structures are described as a function of the phase angles (going from 0° to 360° by 18° increment), for an amplitude of 35, and are numbered from wheel.0 to wheel 19. The coordinate files (pdb format) are downloadable. Each template offers the possibility to generate more […]
Nucleotide 3D Structures around the Conformational Wheel
The structures are described as a function of the phase angles (going from 0° to 360° by 18° increment,), for an amplitude of 35, and are numbered from wheel.0 to wheel.19.(courtesy, Dr. Richard Lavery) wheel.0_0_35 wheel.0_0_35 wheel.1_18_35 wheel.1_18_35 wheel.0_0_35 wheel.0_0_35 wheel.1_18_35 wheel.1_18_35 wheel.4_72_35 wheel.4_72_35 wheel.5_90_35 wheel.5_90_35 wheel.6_108_35 wheel.6_108_35 wheel.7_126_35 wheel.7_126_35 wheel.8_144_35 wheel.8_144_35 wheel.9_162_35 wheel.9_162_35 wheel.10_180_35 […]
Presentation
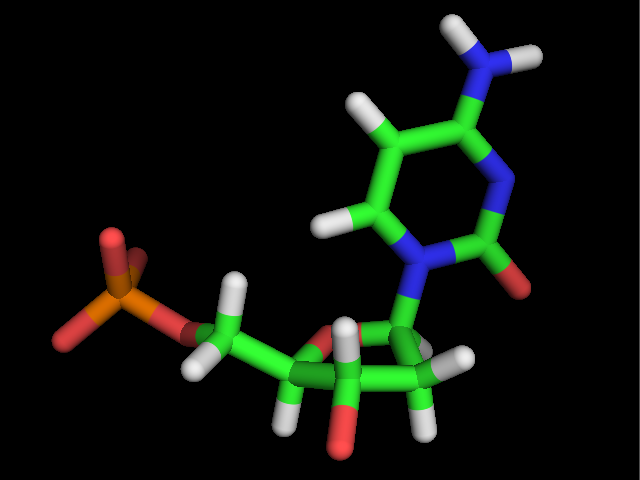
This chapter offers a library of 150 monosaccharides structures found as components of bio-actives glycans, oligo and polysaccharides. They are presented throughout several levels of structural depiction, i.e. one dimension, two-dimension and three-dimension. The monosaccharides are listed in six distinct chapters following alphabetic order ; Complementary to its 3-Dimensional structure, each monosaccharide is presented in the form […]
Abequose …….. Arabinose
Abequose α-D Abequose α-D Abequose α-D Abequose α-D Abequose α-D AceA α-L (Northern conformation) AceA α-L (Northern conformation) AceA α-L (Northern conformation) AceA α-L (Northern conformation) AceA α-L (Northern conformation) AceA α-L (Southern conformation) AceA α-L (Southern conformation) AceA α-L (Southern conformation) AceA α-L (Southern conformation) AceA α-L (Southern conformation) AceA β-L (Northern conformation) AceA […]
Bradyrhizose …… Fucose
α-D α-L β-D β-L Bradyrhizose Bradyrhizose Bradyrhizose Bradyrhizose Bradyrhizose Caryophyllose ; 3,6,10-trideoxy-4-C-[(R)-1-hydroxyethyl]-d-erythro-d-gulo-decose Caryophyllose Caryophyllose Caryophyllose Caryophyllose DAG α-D : Di-Amino α-D Glucose DAG α-D : Di-Amino α-D Glucose DAG α-D : Di-Amino α-D Glucose DAG α-D : Di-Amino α-D Glucose DAG α-D : Di-Amino α-D Glucose DAG β-D : Di-Amino β-D Glucose DAG β-D : […]
Galactose
Galactopyranose α-D Galactopyranose α-D Galactopyranose α-D Galactopyranose α-D Galactopyranose α-D Galactopyranose α-L Galactopyranose α-L Galactopyranose α-L Galactopyranose α-L Galactopyranose α-L Galactopyranose 3,6 anhydro α-D Galactopyranose 3,6 anhydro α-D Galactopyranose 3,6 anhydro α-D Galactopyranose 3,6 anhydro α-D Galactopyranose 3,6 anhydro α-D Galactopyranose 3,6 anhydro α-L Galactopyranose 3,6 anhydro α-L Galactopyranose 3,6 anhydro α-L Galactopyranose 3,6 anhydro α-L […]
Glucose
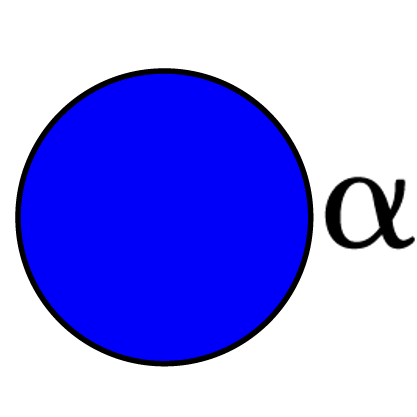
Glucopyranose α-D Glucopyranose α-D Glucopyranose α-D Glucopyranose α-D Glucopyranose α-D Glucopyranose 2,3-S α-D Glucopyranose 2,3-S α-D Glucopyranose 2,3-S α-D Glucopyranose 2,3-S α-D Glucopyranose 2,3-S α-D Glucopyranose 2,3,6-S α-D Glucopyranose 2,3,6-S α-D Glucopyranose 2,3,6-S α-D Glucopyranose 2,3,6-S α-D Glucopyranose 2,3,6-S α-D Glucopyranose 2,6-S α-D Glucopyranose 2,6-S α-D Glucopyranose 2,6-S α-D Glucopyranose 2,6-S α-D Glucopyranose 2,6-S α-D […]
Guluronic ………..MurNAc
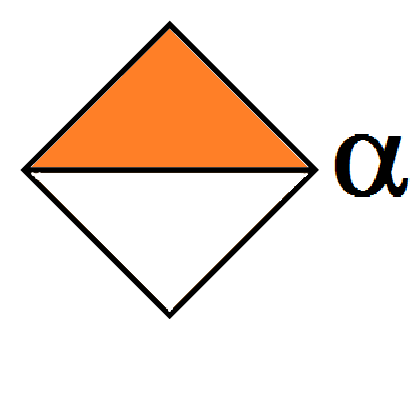
Gulopyranuronic α-D Gulopyranuronic α-D Gulopyranuronic α-D Gulopyranuronic α-D Gulopyranuronic α-D Gulopyranuronic α-L Gulopyranuronic α-L Gulopyranuronic α-L Gulopyranuronic α-L Gulopyranuronic α-L D-Heptose α-D D-Heptose α-D D-Heptose α-D D-Heptose α-D D-Heptose α-D Heptose 6 deoxy Heptose 6 deoxy Heptose 6 deoxy Heptose 6 deoxy Heptose 6 deoxy Idopyranuronic α-L (1C4 conf.) 20a. Idopyranuronic α-L (1C4 conf.) 20b. […]
Neuraminic Acid… Xylose
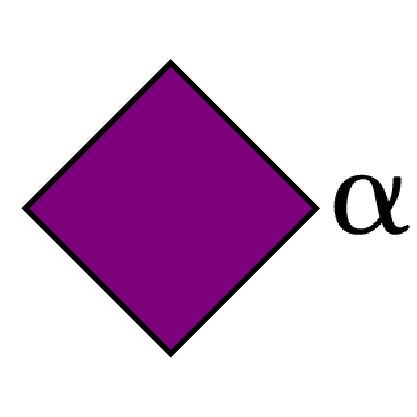
N-Acetyl Neuraminic Acid α-D N-Acetyl Neuraminic Acid α-D N-Acetyl Neuraminic Acid α-D N-Acetyl Neuraminic Acid α-D N-Acetyl Neuraminic Acid α-D N-Glycolyl Neuraminic Acid α-D N-Glycolyl Neuraminic Acid α-D N-Glycolyl Neuraminic Acid α-D N-Glycolyl Neuraminic Acid α-D N-Glycolyl Neuraminic Acid α-D Parp α-D Parp α-L Quinovose α-D Quinovose α-D Quinovose α-D Quinovose α-D Quinovose α-D Quinovose […]
Conventions for Pictorial Representation
The representation of monosaccharides can be concatenated using a limited number of structural descriptors to achieve a fairly exhaustive nomenclature which can be further used in constructing three dimensional structures of glycans. The conventions used for the pictorial/pictorial representations of monosaccharides are the following. Residue Letter Name: Rib, Ara, Xyl, Lyx, All, Alt, Glc, Man, […]
Programme

From Monosaccharides to Polysaccharides, from Structures to 3D Databases (S. Perez) Article: Protein-Carbohydrate interactions: Molecular Modeling Insights Solution Studies : Analytical Ultracentrifugation, SAXS, SANS, MALS Documentation for Practicals (Ch. Ebel) Protein Crystallography and Kinetic Crystallography : A Case Study with a Glycosyl Transferase (A. Royant) Application of Neutron Diffraction to Glycoscience (T. Forsyth) Protein-Sugar Interactions: […]
Structural Glycoscience Grenoble 2018: Program & Documents
From Monosaccharides to Polysaccharides, from Structures to 3D Databases. > http://glycopedia.eu/IMG/pdf/grenoble2018_perez.pdf. Serge Perez Docking and Molecular Modelling. >http://glycopedia.eu/IMG/pdf/grenoble2018_fadda.pdf]. Elisa Fadda Protein-sugar Interaction Isothermal Titration Calorimetry. Bruce Turnbull Application of Neutron Diffraction to Glycoscience. Protein-sugar complexes by X-ray crystallography. >http://glycopedia.eu/IMG/pdf/grenobe2018_varrot.pdf] Annabelle Varrot Solution studies: Analytical ultracentrifugation, SAXS, SANS, MALS. >http://glycopedia.eu/IMG/pdf/grenoble2018-ebel.pdf]. Christine Ebel Protein-Sugar Interaction by Surface […]
Glyco Course Firenze 2016
A World of Sugars a_world_of_sugars_firenze_2016.pdf Reinventing Chemistry whitesides-2015-angewandte_chemie_international_edition.pdf Integrating Knowledge integrating_knowledge_2016.pdf Synchrotron Sources synchrotron_sources_firenze_sp__handout.pdf Herculaneum papyri revealing_letters_in_rolled_herculaneum.pdf
Potential Energy Surfaces
Araf b1-3 Rha pdf/araf_b1-3_rhap.pdfFuc a1-2 Gal pdf/fuc_a1-2_gal.pdfFuc a1-2 Glc pdf/fuc_a1-2_glc.pdfFuc a1-3 Gal pdf/fuc_a1-3_gal.pdfFuc a1-3 Glc pdf/fuc_a1-3_glc.pdfFuc a1-3 GlcNAc pdf/fuc_a1-3_glcnac.pdfFuc a1-4 Gal pdf/fuc_a1-4_gal.pdfFuc a1-4 Glc pdf/fuc_a1-4_gal-2.pdfFuc a1-4 GlcNAc pdf/fuc_a1-4_glc.pdfFuc a1-6 GlcNAc pdf/fuc_a1-6_glcnac.pdfGal a1-2 Fuc pdf/galp_a1-2_fucp.pdfGal a1-2 Gal pdf/gal_a1-2_gal.pdfGal a1-3 Gal pdf/gal_a1-3_gal.pdfGal a1-3 Galf pdf/gal_a1-3_galf.pdfGal a1-3 GalNAcpdf/gal_a1-3-galnac.pdfGal a1-3 Glc pdf/gal_a1-3_glc.pdfGal a1-3 Rha pdf/gal_a1-3_rha.pdfGal a1-4 Gal pdf/gal_a1-4_gal.pdfGal a1-4 GlcNAc pdf/gal_a1-4_glcnac.pdfGal a1-6 Gal pdf/gal_a1-6_gal.pdfGal a1-6 Glc pdf/gal_a1-6_glc.pdfGal b1-2 Gal pdf/gal_b1-2_gal.pdfGal b1-2 Xyl pdf/gal_b1-2_xyl.pdfGal b1-3 Gal pdf/gal_b1-3-gal.pdfGal b1-3 GalNAc pdf/gal_b1-3-galnac.pdfGal b1-3 Glc pdf/gal_b1-3_glc.pdfGal b1-3 GlcNAc pdf/gal_b1-3_glcnac.pdfGal […]
499 Road Map Glycosciences Europe
a-roadmap-for-glycoscience-in-europe_rts_comments_june_2019.pdf Documents joints
Glycans Visual Models
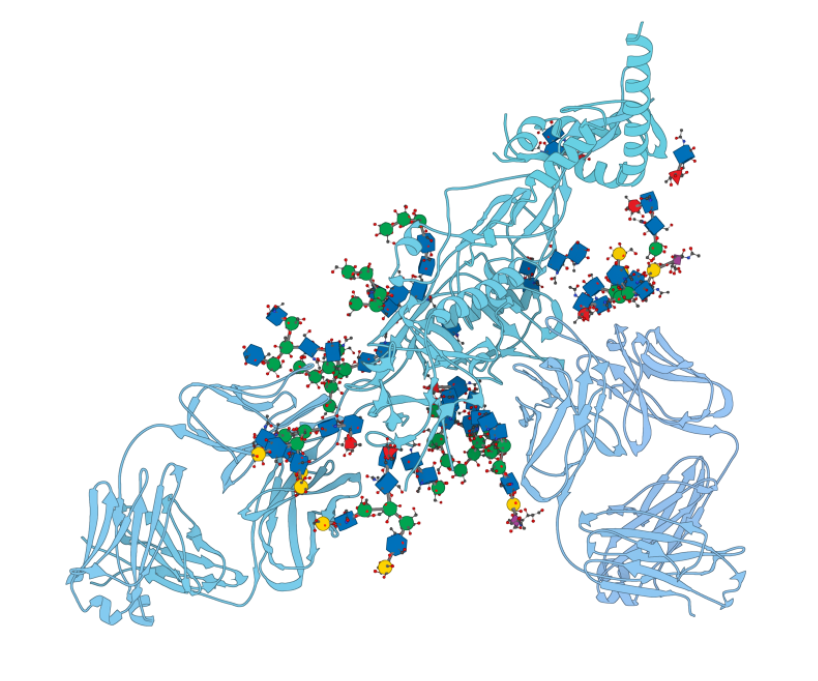
Visualize glycans with 3D Symbols and make cloud simulations. https://www.samson-connect.net/element/df5fa444-ccba-499d-34d7-226132340a25.html This extension provides a visual model for glycans corresponding to Symbol Nomenclature for Glycans (SNFG) in 3D (https://www.ncbi.nlm.nih.gov/glycans/snfg.html). Press Ctrl+Shift+V or go to the Visualization menu > Visualize > Visual model to apply this visual model. Then, in the pop-up dialog, choose “Glycans” from the […]
Glyco 2030 Document
glyco_2030_a_roadmap_for_glycoscience_in_europe.pdf Documents joints
CADD_UGA_October 2018
cadd_novembre2018-2.pdf Documents joints
Glycoscience@synchrotron
glycoscience_synchrotron.pdf Documents joints
Advances_Carbohydrate-Protein Interactions
advances_sp.pdf Documents joints
Computer Aided Drug Design
cadd_novembre2017.pdf Documents joints