The innate immune system functions using at least two recognition strategies: it is capable to distinguish microbial non-self from missing self. These recognition events occur thanks to a range of receptors that all cells of the innate immune system are equipped with, and through which various signals can be triggered by cytokines, conserved components of microorganisms, complement components and antibodies produced by B lymphocytes. The recognition of missing self is based on the recognition of molecules expressed only on healthy uninfected host cells, which leads to inhibition of innate immunity response, and the expression of these molecules is lost once the cells get infected, i.e. the missing self emerges. The missing self recognition plays an important role in the function of NK cells and complement (DeFranco, A, Locksley, R, & Robertson, M. 2007, Paul, W. E. 2008).
The recognition of non-self is based on the recognition of conserved molecular structures that are unique to microorganisms and that are not produced by the host. These structures are called Pathogen Associated Molecular Patterns (PAMPs) and include lipopolysaccharide (LPS) of gram negative (G-) bacteria, lipoteichoic acid (LTA) of gram-positive bacteria (G+), peptidoglycans, lipoproteins generated by palmitoylation of the N-terminal cysteines of many bacterial cell wall proteins, lipoarabinomannan of mycobacteria, double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA) produced by most viruses during the infection cycle, and b-glucans and mannans in fungal cell walls (Paul, W. E. 2008).
1. The diversity of Pattern Recognition Receptors
The receptors of the innate immune system that recognize PAMPs are called Pattern Recognition Receptors (PRRs). The main functions of PRRs include pathogen-induced phagocytosis, activation of pro-inflammatory signaling pathways, opsonization, activation of complement and coagulation cascades, and induction of apoptosis (Janeway, C. A & Medzhitov, R. 2002). All PRRs can be defined as humoral or cell associated, and the latter ones can be subdivided to intracellular and cell surface molecules (fig. 4).The selected examples of PRRs and their ligands are listed in table 1.
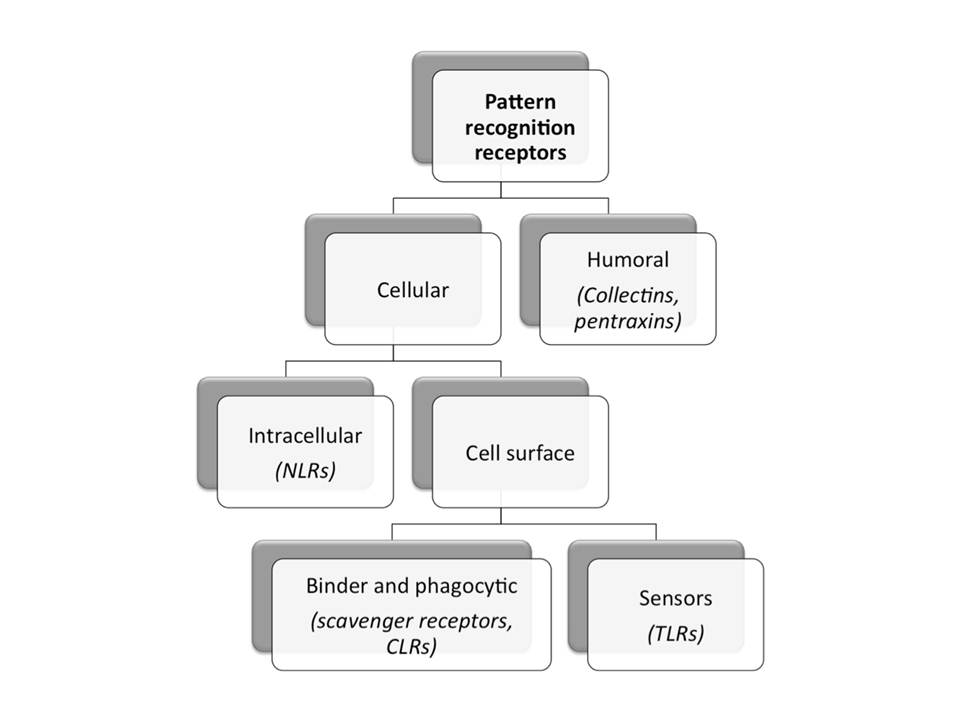
Schematic representation of different groups of pattern recognition receptors with selected examples.
Secreted, or humoral, PRRs may activate complement, opsonize microbial cells to facilitate their phagocytosis, and in some cases function as accessory proteins for PAMP recognition by transmembrane receptors like Toll-like receptors (TLRs). Examples of humoral PRRs include the mannan-binding lectin (MBL), C-reactive protein (CRP), serum amyloid protein (SAP) and peptidoglycan-recognition proteins (PGRPs) (Janeway, C. A & Medzhitov, R. 2002, Kishore, U. 2009).
Cell surface PRRs may be either phagocytic/endocytic or sensor in nature. The sensor receptors do not bind or internalize ligand directly, but recognize PAMPs and induce pro-inflammatory signaling cascades, which lead to various antimicrobial effector responses. They include TLRs, but also many intracellular PRRs are sensing molecules (Kishore, U. 2009). Examples of intracellularly functioning sensors include NOD-like receptors (NLRs), protein kinase R (PKR), 2’-5’-oligoadenylate synthase (OAS)/RNaseL, and retinoid acid–inducible gene–1 (RIG-1)-like receptors (RLRs).
Phagocytic receptors bind and internalize ligands directly in a temperature dependent, saturable and inhibitable ligand binding manner, characteristic to classical receptors. These receptors include scavenger and C-type lectin receptors (CLRs). Scavenger receptors play several roles: they are important in uptake and clearance of degenerate components, such as modified host molecules and apoptotic cells, and they bind and internalize microorganisms and their products (Peiser, L, Mukhopadhyay, S, & Gordon, S. 2002). CLRs are a diverse family of receptors with the ability to bind to carbohydrate moieties and with variable physiological functions from cell adhesion to pattern recognition (Lombardi, G & Riffo-Vasquez, Y. 2008). The cellular and humoral arms of the innate immune system collaborate and maintain host defense.
Examples of selected PRRs and their ligands
Abbreviations: Ac-LDL, acetylated low density lipoprotein; AGE, advanced glycation end-product; DAP, diaminopimelic acid; GM-tripeptide, N-acetyl-D-glucosaminyl-b(1,4)-N-acetylmuramyl-L-Ala-D-Glu; GPI, Glycosylphosphatidylinositol; LDL, Low-Density Lipoproteins; LTA, lipoteichoic acid; MØ, macrophages; MDP, muramyl dipeptide; Mo, monocytes; Neu, neutrophils; Ox-LDL, oxidized LDL; UGRP-1, Uteroglobin-Related Protein 1. Information collected from [11, 12, 15, 16].
Receptor family : TLRS
| Localization | Cell types | Receptor | Ligands | Ligand origin |
| Ubiquitous | Ubiquitous | TLR1 | Triacyllipopeptides | llipopeptides |
| Cell surface | Myeloid cells;mast cells; NKs; DCs; αβ and γδ T cells | TLR2 | Envelope proteins GPI-linked proteins Lipoproteins LTA Peptidoglycans Zymosan | Virus Trypanosomes Mycobacteria G+ bacteria G+ bacteria Fungi |
| Intracellular (endosomal) | DCs; NKs | TLR3 | dsRNA | Viruses |
| Cell surface | Mo; mast cells; Neu; γδ T cells; Golgi in gut epithelial cells | TLR4 | Fusion protein Glycoinositol phospholipids LPS Mannan | Respiratory syncytial virus Fungi G+ bacteria Fungi |
| Cell surface | Epithelial cells;NKs; Mo; DCs | TLR5 | Flagellin | Bacteria |
| Cell surface | Myeloid cells; mast cells; B cells | TLR6 | Diacyl lipopeptides LTA Zymosan | Mycobacteria G+ bacteria Fungi |
| Intracellular (endosomal) | DCs; B cells; Eosinophils | TLR7 | ssRNA | Viruses |
| Intracellular (endosomal) | NKs; T cells; myeloid cells | TLR8 | ssRNA | Viruses |
| Intracellular (endosomal) | DCs; B cells; surface of tonsillar cells | TLR9 | CpG-containing DNA Herpes virus DNA | Bacteria, protozoa, virus Viruses |
| Cell surface | DCs; B cells | TLR10 | Unknown | Unknown |
Examples of selected PRRs and their ligands.
Abbreviations: Ac-LDL, acetylated low density lipoprotein; AGE, advanced glycation end-product; DAP, diaminopimelic acid; GM-tripeptide, N-acetyl-D-glucosaminyl-b(1,4)-N-acetylmuramyl-L-Ala-D-Glu; GPI, Glycosylphosphatidylinositol; LDL, Low-Density Lipoproteins; LTA, lipoteichoic acid; MØ, macrophages; MDP, muramyl dipeptide; Mo, monocytes; Neu, neutrophils; Ox-LDL, oxidized LDL; UGRP-1, Uteroglobin-Related Protein 1. Information collected from [11, 12, 15, 16].
Receptor family : Scavenger Receptors
| Localization | Cell types | Receptor | Ligands | Ligand origin |
| Cell Surface | MØ; DCs; Certain endothelial cells | SR-A | LPS and LTA Unidentified protein ligand in serum; activated B cells β amyloid protein; apoptotic cells; Ox-LDL and Ac-LDL; AGE modified proteins | Microbial cell walls Endogenous Modified self |
| Cell Surface | MØ; DCs; Certain endothelial cells | MARCO | LPS UGRP-1 Ac-LDL | Microbial cell walls Endogenous Modified self |
| Cell Surface | MØ; DCs; Certain endothelial cells | SRCL-1 | G+, G- bacteria, yeast T and Tn antigen Ox-LDL | Microbes Endogenous Modified self |
Examples of selected PRRs and their ligands.
Abbreviations: Ac-LDL, acetylated low density lipoprotein; AGE, advanced glycation end-product; DAP, diaminopimelic acid; GM-tripeptide, N-acetyl-D-glucosaminyl-b(1,4)-N-acetylmuramyl-L-Ala-D-Glu; GPI, Glycosylphosphatidylinositol; LDL, Low-Density Lipoproteins; LTA, lipoteichoic acid; MØ, macrophages; MDP, muramyl dipeptide; Mo, monocytes; Neu, neutrophils; Ox-LDL, oxidized LDL; UGRP-1, Uteroglobin-Related Protein 1. Information collected from [11, 12, 15, 16].
Receptor family : Collectins
| Localization | Cell types | Receptor | Ligands | Ligand origin |
| Humoral | MBL | Terminal mannose Residues | Bacterial surfaces | |
| Localization | Cell types | Receptor | Ligands | Ligand origin |
| Humoral | CRP | Phosphorylcholine | Bacterial surfaces | |
| Humoral | SAP | Phosphorylcholine | Bacterial surfaces |
Examples of selected PRRs and their ligands.
Abbreviations: Ac-LDL, acetylated low density lipoprotein; AGE, advanced glycation end-product; DAP, diaminopimelic acid; GM-tripeptide, N-acetyl-D-glucosaminyl-b(1,4)-N-acetylmuramyl-L-Ala-D-Glu; GPI, Glycosylphosphatidylinositol; LDL, Low-Density Lipoproteins; LTA, lipoteichoic acid; MØ, macrophages; MDP, muramyl dipeptide; Mo, monocytes; Neu, neutrophils; Ox-LDL, oxidized LDL; UGRP-1, Uteroglobin-Related Protein 1. Information collected from [11, 12, 15, 16].
Receptor family : NLRS
| Localization | Cell types | Receptor | Ligands | Ligand origin |
| INTRACELLULAR | Lymphocytes, MØ, DCs, epithelial and mesothelial cells | Nod1 | GM-tripeptide meso-lanthionine, meso-DAP γ-D-Glu-DAP D-lactyl-L-Ala-γ-Glumeso- DAP-Gly heptanolyl-γ-Glumeso-DAP-Ala | Helicobacter pylori Shigella flexneri Listeria monocytogenes Campylobacter jejuni Enteropathogenic Escherichia coli Chlamydia pneumoniae Pseudomonas aeruginosa Bacillus spp |
| INTRACELLULAR | Lymphocytes, MØ, DCs, epithelial and mesothelial cells | Nod2 | MDP MurNAc-L-Ala-γ-DGlu-L-Lys | Streptococcus pneumoniae Listeria monocytogenes Mycobacterium tuberculosis Salmonella typhimurium Staphylococcus aureus Shigella flexneri |
| INTRACELLULAR | Lymphocytes, MØ, DCs, epithelial and mesothelial cells | Nlrc4 | flagelin | Salmonella typhimurium Legionella pneumoniae Pseudomonas aeruginosa |
| INTRACELLULAR | Lymphocytes, MØ, DCs, epithelial and mesothelial cells | Nlrp1b | anthrax lethal toxin | Bacillus anthracis |
| Lymphocytes | Bacterial and viral | |||
| INTRACELLULAR | Lymphocytes, MØ, DCs, epithelial and mesothelial cells | Nlrp3 | RNA, viral DNA, uric acid crystals, LPS, LTA, MDP, silica, asbestos |
2. C-type lectin receptors as PRRs
The C-type lectin family comprises a large group of Metazoan proteins that contain C-type lectinlike domains (CTLDs). Although originally CTLDs were identified as the structures that bind carbohydrates in a Ca2+-dependent manner (thereof the term C-type), not all the members of this family recognize carbohydrates and not all need Ca2+ for ligand binding. Therefore, CTLDs are referred as “C-type lectin-like domains” (Zelensky, A. N & Gready, J. E. 2005).
The mammalian C-type lectin receptors (CLRs) are divided into 17 types based on their phylogenetic relationships and domain structures (fig. 5). Most of the CLR family members function as adhesion receptors, and only CLRs of type II, V and VI are present mostly on myeloid lineage immune cells and function as PRRs, while type III CLRs are soluble PRRs (Geijtenbeek, T. B. H & Gringhuis, S. I. 2009).
![The structural diversity of CLR family.
CLR types are marked by roman numbers : I – lecticans, II – asialoglycoprotein receptor (ASGR) DC receptor group,III – collectins, IV – selectins, V – NK receptors, VI – multi-CTLD endocytic receptors (macrophage mannose receptor group), VII – Reg proteins, VIII – chondrolectin group, IX – tetranectin group, X – polycystin 1, XI – attractin, XII – EMBP (eosinophil major basic protein), XIII –DGCR2 (the product of DiGeorge syndrome critical region gene 2), XIV – thrombomodulin group, XV – Bimlec, XVI – SEEC (soluble protein containing SCP, EGF, EGF and CTLD domains), XVII – CBCP (Calx-b and CTLD containing protein). (Adapted from [Zelensky, A. N & Gready, J. E. (2005)])](https://glycopedia.eu/wp-content/uploads/2013/03/fig.5-cc9.jpg)
CLR types are marked by roman numbers : I – lecticans, II – asialoglycoprotein receptor (ASGR) DC receptor group,III – collectins, IV – selectins, V – NK receptors, VI – multi-CTLD endocytic receptors (macrophage mannose receptor group), VII – Reg proteins, VIII – chondrolectin group, IX – tetranectin group, X – polycystin 1, XI – attractin, XII – EMBP (eosinophil major basic protein), XIII –DGCR2 (the product of DiGeorge syndrome critical region gene 2), XIV – thrombomodulin group, XV – Bimlec, XVI – SEEC (soluble protein containing SCP, EGF, EGF and CTLD domains), XVII – CBCP (Calx-b and CTLD containing protein). (Adapted from [Zelensky, A. N & Gready, J. E. (2005)])
CLRs that function as PRRs are mostly expressed by different DC subsets (table 2). They bind pathogens through the recognition of mannose, fucose, glucan and other carbohydrate structures. The combination of CLRs on DCs enables the recognition of most classes of human pathogens. Pathogen recognition by CLRs leads to its internalization, degradation and subsequent antigen presentation (Geijtenbeek, T. B. H & Gringhuis, S. I. 2009).
Selected examples of CLRs that function as PRRs
Type II (Ca2+-dependent CRD)
| CLR | Expression | Recognized glycans | Glycans from pthogen | Endogenous ligands |
| DC-SIGN (CLEC4L, CD209) | myDCs | High mannose and fucose (LewisX, LewisY, LewisA, LewisB) | Bacteria: Mycobacterium tuberculosis; Mycobacterium leprae; BCG; Lactobacilli spp.; Streptococcus pneumoniae; Leptospira interrogans; Helicobacter pylori Viruses: HIV-1; measles virus; Dengue virus; HCV; CMV; SARS coronavirus; HSV; H5N1; WNV; Ebola virus and other filoviruses; phlebo viruses Fungi: Candida albicans; Aspergillus fumigatus Protozoa: Leishmania spp. Other: Tick Ixodes capularis saliva protein Salp15; peanut allergen Ara h1; Schistosoma mansoni soluble egg antigens | ICAM-2; ICAM-3; CEA |
| DC-SIGNR (CLEC4M,CD299) | Endothelial cells in lymph-node sinuses; liver sinusoidal endothelial cells | Mannosylated glycans, high mannose N-glycans | HIV-1; Ebola; Schistosoma mansoni | ICAM-1; ICAM-2; ICAM-3 |
| Langerin (CLEC4K, CD207) | LCs; dermal DC subset | High mannose, fucose (LewisY, LewisB), GlcNAc, β-glucans, sulphated sugars (heparin) | Bacteria: Mycobacterium leprae Fungi: Candida; Saccharomyces cerevisiae; Malassezia furfur Viruses: HSV; Measles virus; HIV-1 | Type I pro-collagen |
| MGL (CLEC10A, CD301) | myDCs; MØ | Terminal GalNAc (Tn, LDN and LDNF antigens) | Filoviruses; Influenza virus; Schistosoma mansoni | CD45; gangliosides; MUC-1 |
| Dectin-2 (CLEC6A) | myDCs; pDCs; Mo; MØ; B cells; Neu | High mannose | Bacteria: Mycobacterium Tuberculosis Fungi: Candida albicans; Trichophyton rubrum; Aspergillus fumigatus; Microsporum audounii; Paracoccoides brasiliensis Allergens: House dust mite Dermatophagoides pteronyssinus allergens | Unknown |
| Mincle (CLEC4E) | myDCs; Mo; MØ | a-mannose | Malassezia spp.; Mycobacteria; Candida | Damaged cells |
| DCIR (CLEC4A) | myDCs; pDCs; Mo; MØ; B cells; Neu | Unknown | HIV-1 | Unknown |
| BDCA2 (CLEC4C, CD303) | pDCs; Mo; MØ; Neu | Unknown | Unknown | Unknown |
Selected examples of CLRs that function as PRRs.
V (Ca2+-independent CRD)
| CLR | Expression | Recognized glycans | Glycans from pthogen | Endogenous ligands |
| Dectin-1(CLEC7A) | myDCs; Mo; MØ; B cells | β-1,3-glucan | Bacteria: Mycobacterium tuberculosis Mycobacterium abscessus Fungi: Candida albicans; Aspergillus fumigatus; Pneumocystis carinii; Penicillium marneffei; Coccidioides posadasii; Histoplasma capsulatum | Ligand on T cells |
| MICL (CLEC12A) | myDCs; Mo; MØ; Neu | Unknown | Unknown | Unknown |
| CLEC2 (CLEC1B) | Platelets, peripheral blood Neu | Unknown | HIV-1; Snake venom rhodocytin; podoplanin | Unknown |
The CLRs can be immune activating or inhibitory depending on their ability to associate with certain signaling molecules or the presence of specific motifs in their cytoplasmic tails. Most of the type II CLRs are predicted to be activating as in their trans-membrane regions they have a positively charged residue which allows association with adaptor proteins. The activating CLRs may harbour the immunoreceptor tyrosine-based activation motifs (ITAMs), which constitute two YxxI/L motifs separated by 6-12 amino acid spacers (YxxI/Lx(6-12)YxxI/L). Upon ligand binding, clustering of CLRs occurs and ITAMs are phosphorylated, which initiates a downstream signaling cascade eventually leading to activation of various cellular responses. Besides, there are CLRs that bear ITAM-like motifs (N-terminal tyrosin in YxxxL/I) in their cytoplasmic tail, for ex. dectin-1. The activating CLRs include dectin-2, DCAR, BDCA2, Mincle and DC-SIGN [Redelinghuys, P & Brown, G. D. (2011)].
The inhibitory CLRs themselves possess the immunoreceptor tyrosine-based inhibition motifs (ITIMs: I/V/L/SxYxxI/L/V) in their cytoplasmic tails, and in this case the ligand binding to CLR followed by phosphorylation of ITIM initiates signaling cascades that culminate at the inhibition of cellular activation. The examples of inhibitory CLRs include DCIR and MICL. There also exist CLRs that harbour ITIMs but mediate cellular activation (Redelinghuys, P & Brown, G. D. 2011).
The structure of C-type lectin-like domains. The common feature of all CLRs is that they possess at least one CTLD – a compact globular structure with a characteristic fold designated “C-type lectin-like fold” that is unusual to any other known proteins. For the majority of CLRs that function as PRRs, the CTLDs actually bind sugars, usually in Ca2+-dependent manner, and therefore this domain is commonly called a “carbohydrate recognition domain” (CRD) rather than CTLD.
All CRDs possess a characteristic “double-loop” fold (fig. 6). The whole domain can be regarded as a loop with two flanking α helices (α1 and α2) and two antiparallel b-sheets: N- and C-terminal β strands β1 and β5 constitute the basal β-sheet, and the top β-sheet is formed by strands β2, β3, and β4. The long loop region enters and exits the core domain at the same location, and is involved in Ca2+-dependent carbohydrate binding, and for some CRDs in domain-swapping dimerization. Four highly conserved cysteins form two disulphide bridges at the bases of the loops: C1-C4 bridge links α1 and β5, and C2-C3 bridges β3 strand and a loop upstream the β5 strand. Another highly conserved sequence feature of the CRDs is the “WIGL” motif located within β2 strand and believed to stabilize the core of the domain.
The long loop region among different CRDs varies, and those that possess it are designated“canonical”, while those that lack it are called compact. The presence or absence of a short extension at N-terminus, a β1´-hairpin, further subdivides CRDs to long or short forms, respectively. Two additional cysteins at the beginning of CRDs sequence are characteristic for the long form CRDs. The corresponding disulphide bridge (C0-C0´) stabilizes the β-hairpin.
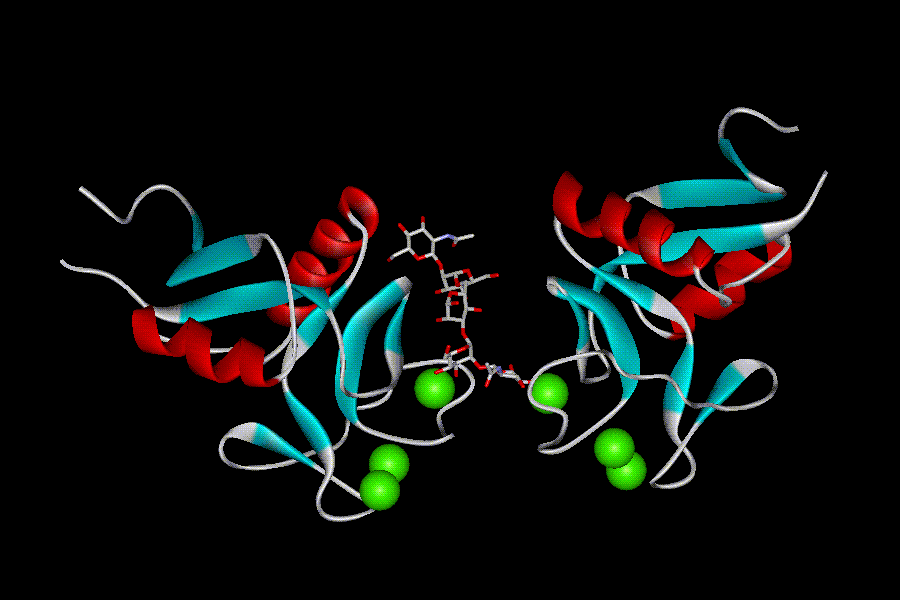
There may be up to four Ca-binding sites in the CRDs, and their occupancy depends on the sequence of a particular CRD and on crystallization conditions. Sites 1, 2, and 3 are located within the long loop, while the fourth site participates in the salt bridge formation between helix a2 and b1/b5 sheet (Zelensky, A. N & Gready, J. E. 2005).
The structural basis of sugar binding within carbohydrate recognition domains. The sugar binding in the CRDs occurs at Ca2+-binding site 2, and both carbonyl sidechains coordinating calcium and Ca2+ itself are involved in sugar binding. Ca2+-coordination at this site is provided by carbonyl sidechains mainly within two characteristic motifs. One of these motifs, EPN or QPD, resides in a long loop region and defines the monosaccharide binding specificity. The second one, a WND motif, is contributed by β4 strand. Additionally, a carbonyl side chain provided by the residue preceding the second conserved cysteine, also participates in Ca2+ coordination at site 2. The schematic representation of Ca2+ coordination and hexose binding is depicted in figure 7A.
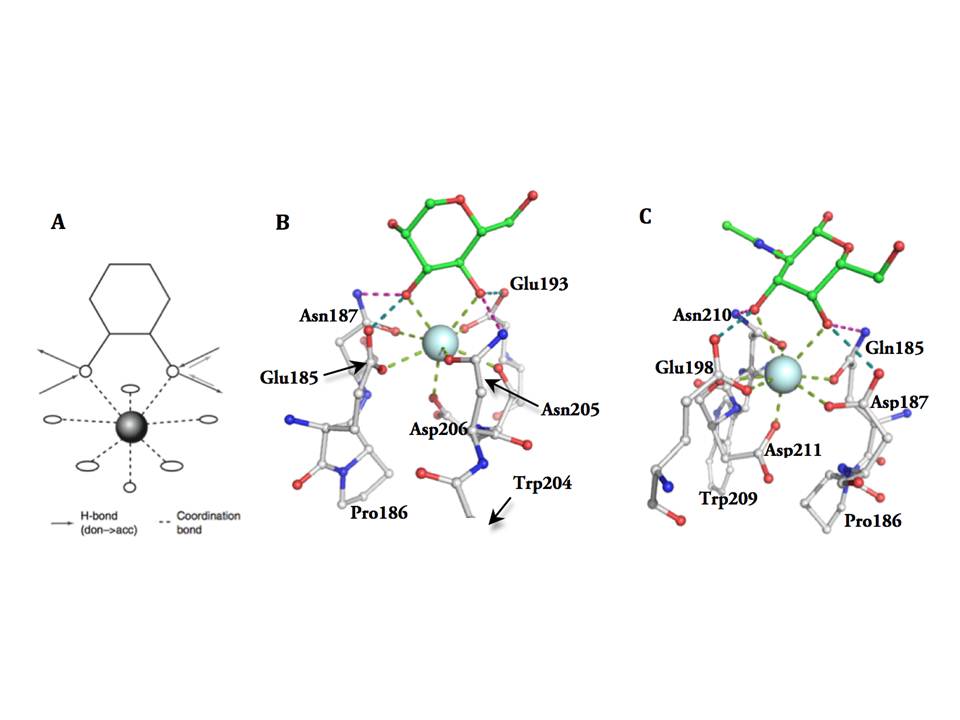
Monosaccharide binding in Ca site 2
The overall network of the hydrogen-bond donors and acceptors in the site determines the binding orientation of the carbohydrate and also which hydroxyls of the carbohydrate it can accept, i.e. the monosaccharide specificity. The EPN motif has a configuration, which accommodates mannose group monosaccharides (fig. 7B), while QPD motif determines specificity for galactose group monosaccharides (fig. 7C). In both of these motifs the cis-configuration of the two carbonyl sidechains separated by proline is crucial for Ca2+ coordination and sugar binding. Besides the H-bond network imposed constraints, other structural elements in the binding sites introduce selectivity to particular ligands within the mannose or galactose groups.
The other three Ca2+ binding sites play the structural stabilization role, as removal of Ca2+ increases susceptibility to proteolysis and changes physical properties of the domain. Ca2+ binding site 2 is also important for structural stability of the domain. It has been shown that pH-induced loss of Ca2+ causes the destabilization of the loops, which has an important physiological role for CRDs of endocytic receptors as internalization of ligand-bound receptor to acidic lysosomes and consequent Ca2+ loss leads to the release of the ligand for further processing, while receptor is recycled to the cell surface (Zelensky, A. N & Gready, J. E. 2005).
