CAZymes (Carbohydrate Active EnZymes) degrade, synthesize, and modify all complex carbohydrates on Earth. CAZymes are extremely important in research on human health, nutrition, gut microbiome, bioenergy, plant disease, and global carbon recycling. Current CAZyme annotation tools are all based on sequence similarity. A more robust approach is to detect protein structural similarity between query proteins and known CAZymes indicative of distant homology. CAZymes3D (https://pro.unl.edu/CAZyme3D/) has been developed to fill the research gap in the lack of dedicated 3D structure databases for CAZymes.
CAZyme3D contains 870,740 AlphaFold predicted 3D structures (the Whole dataset). A subset of CAZyme 3D structures from 188,574 non-redundant sequences (termed the ID50 dataset) were subjected to structural similarity-based clustering analyses. Such clustering allowed the organization of all CAZyme structures using a hierarchical classification that includes existing levels defined by the CAZy database (class, clan, family, subfamily) and newly defined levels (subclasses, structural cluster [SC] groups and SCs).
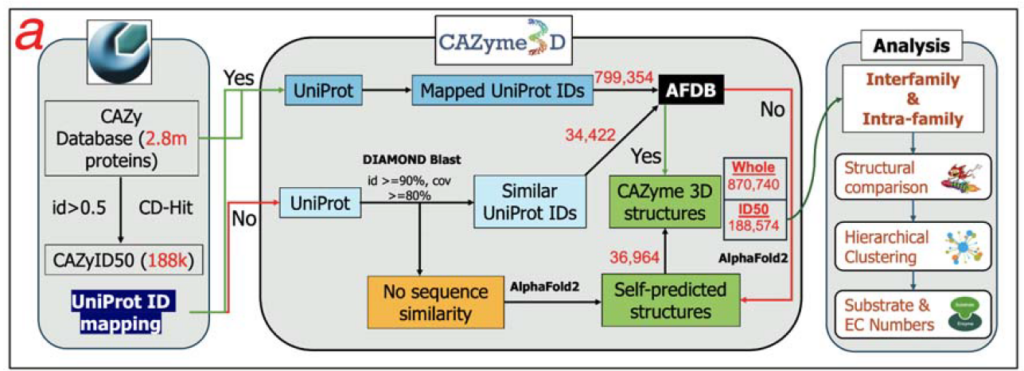
Inter-family structural clustering successfully grouped CAZy families and clans with the same structural folds into the same subclasses. Intra-family structural clustering classified structurally similar CAZymes into SCs, further classified into SC groups. SCs and SC groups differed from sequence similarity-based CAZy subfamilies. Using the CAZyme structures as a search database, the authors created job submission pages where users can submit query protein sequences or PDB structures for a structural similarity search. CAZyme3D will be a valuable new tool to support the discovery of novel CAZymes by providing a comprehensive database of CAZyme 3D structures.




