Enzymatic digestion of lignocellulosic plant biomass is key in bio-refinery approaches for producing biofuels and other valuable chemicals. However, the recalcitrance of this material, in conjunction with its variability and heterogeneity, strongly hampers the economic viability and profitability of biofuel production. To complement academic and industrial experimental research, we designed an advanced web application that encapsulates our in-house developed complex biophysical model of enzymatic plant cell wall degradation. PREDIG (https://predig.cs.hhu.de/) is a user-friendly, free, and fully open-source web application that allows users to perform in silico experiments. Specifically, it uses a Gillespie algorithm to run stochastic simulations of the enzymatic saccharification of a lignocellulose microfibril at the mesoscale in three dimensions. Such simulations can be used to test the action of distinct enzyme cocktails on the substrate.
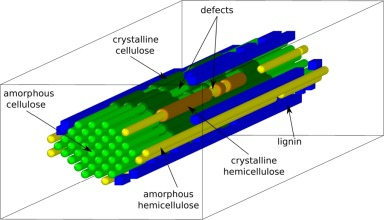
Additionally, PREDIG can fit the model parameters to uploaded experimental time-course data, thereby returning values that are intrinsically difficult to measure experimentally. This allows the user to learn which factors quantitatively explain the recalcitrance to saccharification of their specific biomass material.




