The ultimate microscope, directed at a cell, would reveal the dynamics of all the cell’s components with atomic resolution. In contrast to their real-world counterparts, computational microscopes are currently on the brink of meeting this challenge. In this perspective, the article shows how an integrative approach can be employed to model an entire cell, the minimal cell, JCVI-syn3A, at full complexity.
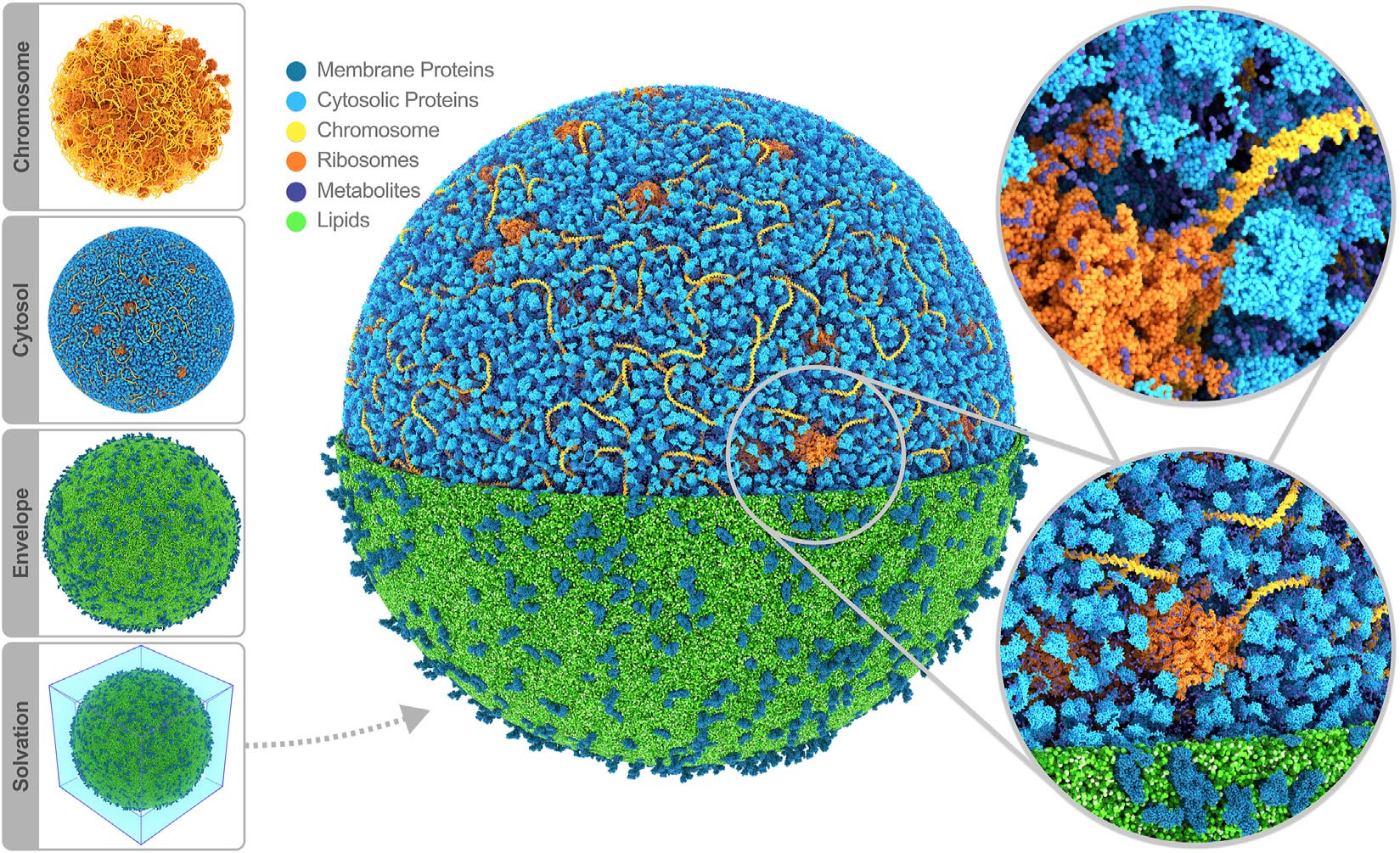
In conclusion, we presented a roadmap toward whole-cell MD simulations, illustrated by constructing the first MD model of an entire cell using our Martini ecosystem. The model represents the next level realized with the computational microscope, providing a complete picture of the cell and directly making it possible to relate molecular structures and interactions to cellular function. In the long term, our computational framework will enable us to study a wide variety of mesoscopic systems, possibly informing the design of fully synthetic cells and modeling cells with more complex internal structures.




