Glycosaminoglycans (GAGs) are informational molecules with astounding structural diversity. Understanding the behaviour of GAGs in the free and protein-bound states is critical for harnessing this diversity. Molecular dynamics (MD) offers atomistic insight into principles governing GAG recognition by proteins. Here, we discuss how MD can be used to understand the local and global properties of GAGs in free solutions, including torsions, puckering, hydrogen bonding, flexibility, and energetics. The authors discuss MD studies on GAG–protein complexes, which help elucidate the strength of interacting residues, the role of water, energetics, and so on.
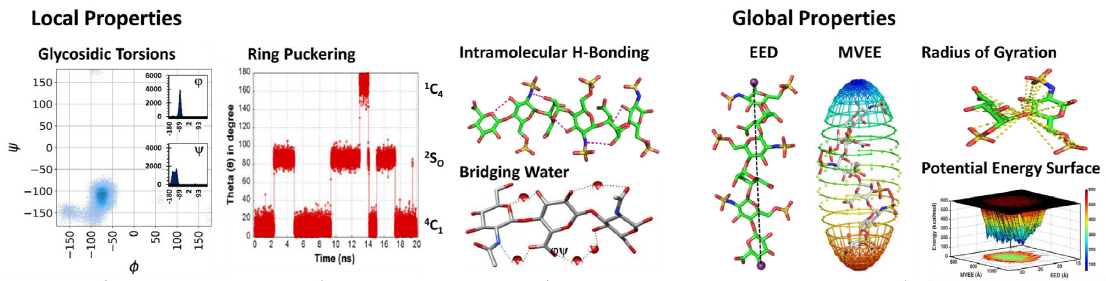
The MD results suggest that GAG recognition of proteins is a continuum from the highly selective end to the fully non-selective on the other with intermediate levels of selectivity, including moderately selective and plastic. The advancements in MD technology, such as coarse-grained MD, coupled with really long simulations will help understand macroscale molecular movements in the future.




