The article describes a new method for designing proteins that bind to a specific site on the surface using no information other than the 3D structure of the target. A general solution to this problem requires a broad exploration of the vast space of possible binding modes to a selected region of a protein surface, followed by an intense search in the vicinity of the most promising binding modes.
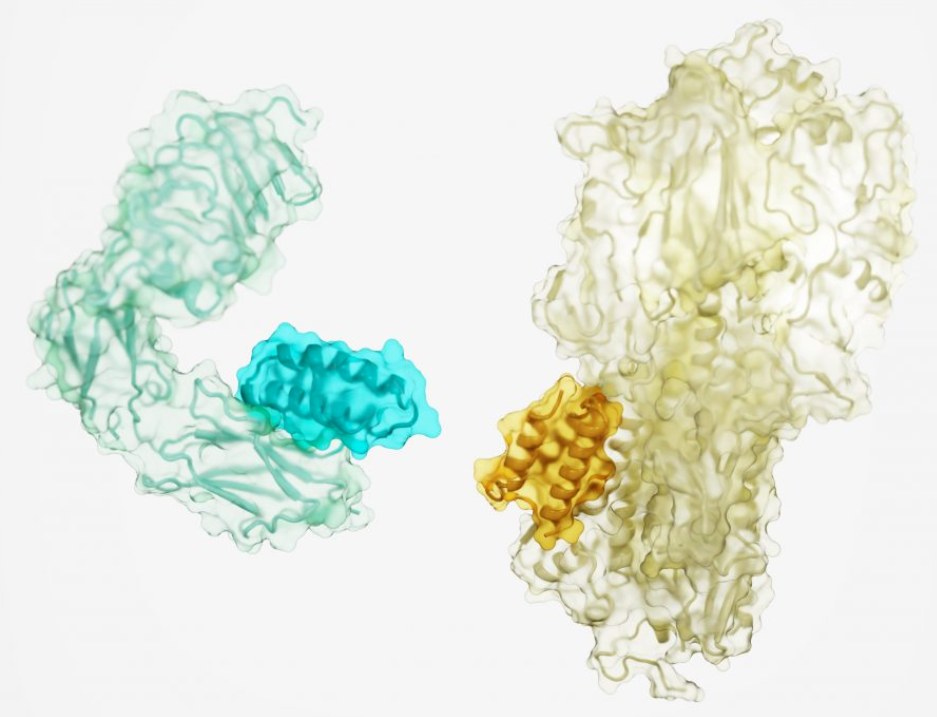
The researchers generated high-affinity binding proteins against 12 distinct molecular targets with different shapes and protein surfaces. These targets include important cellular receptors such as TrkA, EGFR, Tie2, the insulin receptor, and proteins on the surface of the influenza virus and SARS-CoV-2 (the virus that causes COVID-19).
Biophysical characterization shows that the binders are all smaller than 65 amino acids. They are hyper stable, and following experimental optimization, they bind their targets with nanomolar to picomolar affinities. Crystal structures of five of the binder-target complexes were solved; all five are very close to the corresponding computational design models. The methodological achievement solves a long-standing challenge in drug development and may lead to new treatments for cancer, diabetes, infection, inflammation, and beyond.




