Tools to study glycans are rapidly evolving; however, most of the present knowledge is deeply dependent on binding by glycan-binding proteins (e.g., lectins). The specificities of lectins have not always been well-defined, making it difficult to leverage their full potential for glycan analysis. The authors use a combination of machine learning algorithms and expert annotation to define lectin specificity for this important probe set. The investigation uses comprehensive glycan microarray analysis of commercially available lectins, obtained using version 5.0 of the Consortium for Functional Glycomics glycan microarray (CFGv5, made public in 2011).
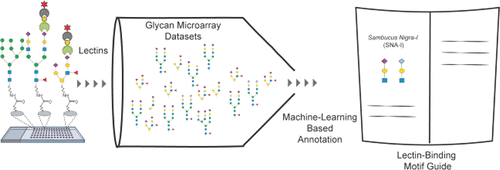
The authors report the creation of this data set and its use in the large-scale evaluation of lectin−glycan binding behaviors. The motif analysis was performed by integrating 68 manually defined glycan features with systematic probing of computational rules for significant binding motifs using mono- and disaccharides and linkages. From a combination of machine learning with manual annotation, the authors create a detailed interpretation of glycan-binding specificity for 57 unique lectins, categorized by their major binding motifs: mannose, complex-type N-glycan, O-glycan, fucose, sialic acid and sulfate, GlcNAc and chitin, Gal and LacNAc, and GalNAc.




