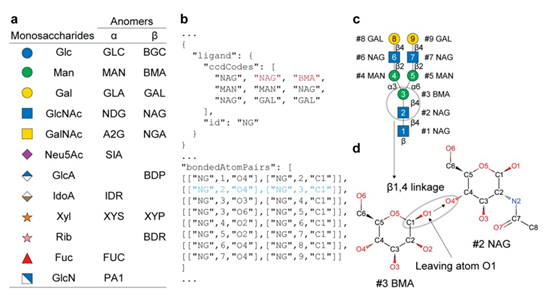
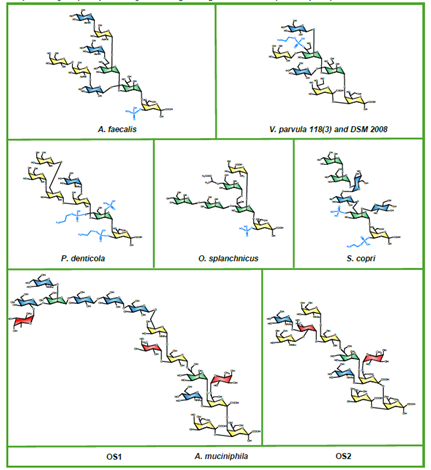
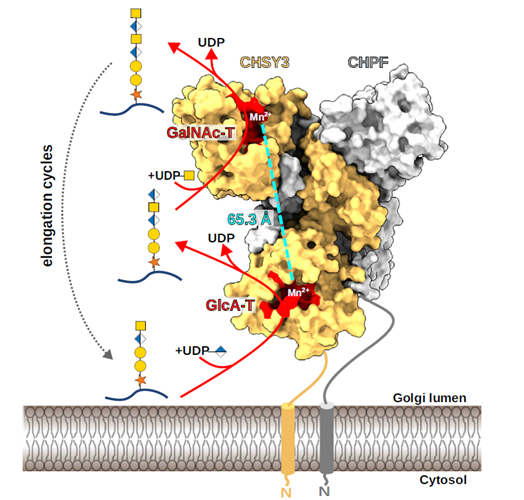
Glycans are complex carbohydrates that exhibit extraordinary structural complexity and stereochemical diversity while playing essential roles in many biological processes, including immune regulation, pathogen recognition, and cell communication. In humans, more than half of all proteins are glycosylated, particularly those in secretory and membrane-associated pathways, highlighting the importance of glycans in health and disease. The recent release of the AlphaFold 3 source code enables customizable modeling not only of proteins but also glycan-containing biomolecular complexes. The authors assessed the capacity of AlphaFold 3 to model glycans using several input formats and identified a hybrid syntax employing Chemical Component Dictionary (CCD)-based molecular building blocks linked by ‘bondedAtomPairs’ (BAP) as most effective in generating stereochemically valid glycan models. This workflow was used to create a library of AlphaFold 3 input templates and corresponding structural models for various glycan classes. Further exploration of capabilities, limitations, and remediation strategies for modeling problematic structures was conducted. Glycan interactions were also modeled with glycosylation enzymes and lectins, with benchmarking and validation against known crystal structures. This protocol-driven approach is valuable for generating stereochemically valid, static models of glycan-protein interactions to support hypothesis development and subsequent structural and functional validation. However, caution should be observed in the overinterpretation of the static models since glycans are known to exhibit considerable conformational dynamics that can be further captured by equilibrium sampling using molecular dynamics-based approaches. By sharing benchmarked examples using the BAP syntax the authors aim to support broader evaluation of AlphaFold 3 in studying glycan- related mechanisms in biosynthesis, signaling, infection, and disease.