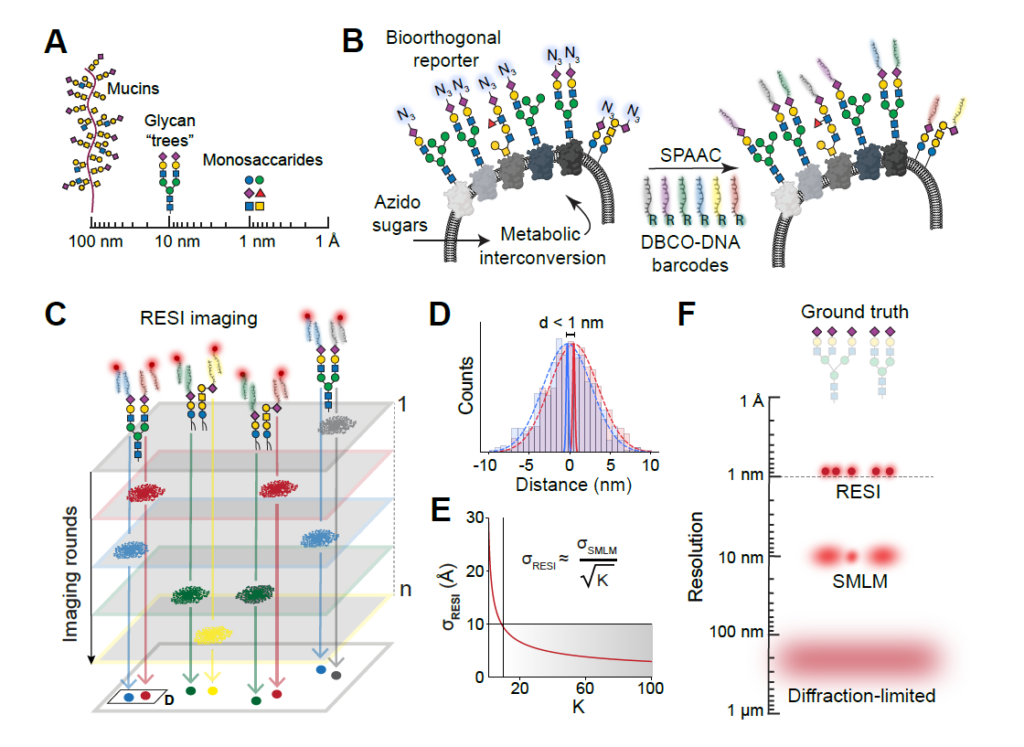
Glycobiology is rooted in the study of monosaccharides, Angstrom-sized molecules that are the building blocks of intricate glycosylation patterns. Glycosylated biomolecules form the glycocalyx, a dense coat encasing every human cell with central relevance – among others – in immunology, oncology, and virology. In order to understand glycosylation function, visualizing its molecular structure is fundamental. However, the ability to visualize the molecular architecture of the glycocalyx has remained elusive. Mass spectrometry, electron microscopy, and fluorescence microscopy lack cellular context, specificity, and resolution. Here, we address these limitations by combining metabolic labeling with Angstrom-resolution fluorescence microscopy, enabling the first-ever visualization of individual sugars within glycans on the cell surface. The present work provides unprecedented insights into the molecular architecture of the glycocalyx and constitutes the foundation for future explorations of its function in health and disease