Carbohydrate-binding modules (CBMs) are discrete protein helper modules with a non-catalytic carbohydrate-binding function that exhibit various binding specificities. CBMcarb-DB is a curated database that classifies the three-dimensional structures of CBM-carbohydrate complexes determined by single crystal X-ray diffraction and solution NMR spectroscopy.
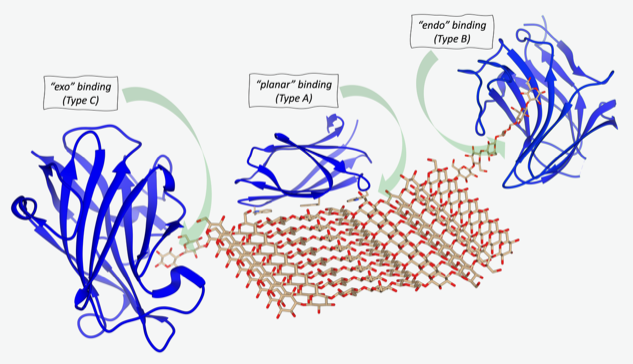
/The database architecture and navigation tools have been designed to query the database using the Protein Data Bank (PDB), UniProtKB and GlyTouCan (universal glycan repository) identifiers. Special attention has been given to describing the bound glycans in a simple graphical representation and numerical format for cross-referencing with other glycoscience and functional data databases.
CBMcarb-DB provides detailed information on CBMs and their bound oligosaccharides and shows their interactions using several open-access applications. It gives examples of how the curated information provided by CBMcarb-DB can be integrated with AI 3D structure prediction algorithms to facilitate structure-function studies. The article highlights the exciting convergence of CBMcarb-DB with the glycan array repository, which serves as a valuable resource for the study of interactions between glycans and various biomolecular targets .CBMcarb-DB is an open resource available at https://cbmdb.glycopedia.eu/ and https://cbmcarb. webhost.fct.unl.pt.