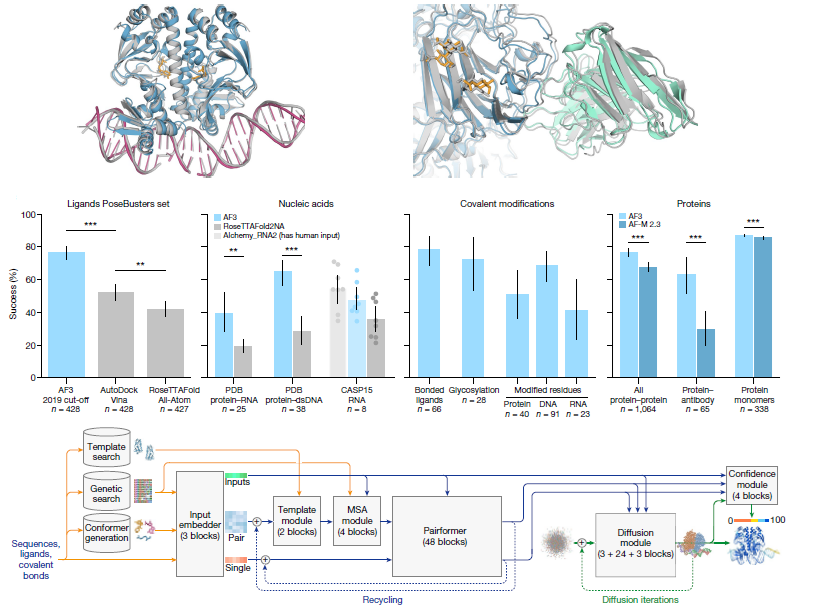
The introduction of AlphaFold 2 has sparked a revolution in the modelling of protein structure and of proteins and their interactions. It enables a wide range of applications in protein modelling and design. The article presents the AlphaFold 3 model with a significantly updated diffusion-based architecture. It is capable of predicting the joint structure of complexes including proteins, nucleic acids, small molecules, ions and modified residues. The new AlphaFold model demonstrates significantly improved accuracy over many previous specialized tools. There is a far greater accuracy for ligand interactions compared to state-of-the-art docking tools, much higher accuracy for protein-nucleic acid interactions compared to nucleic acid-specific predictors. There is a much higher accuracy for antibody-antigen predictions compared to AlphaFold Multimer. Together, these results demonstrate highly accurate biomolecular space within a single, unified deep learning framework.