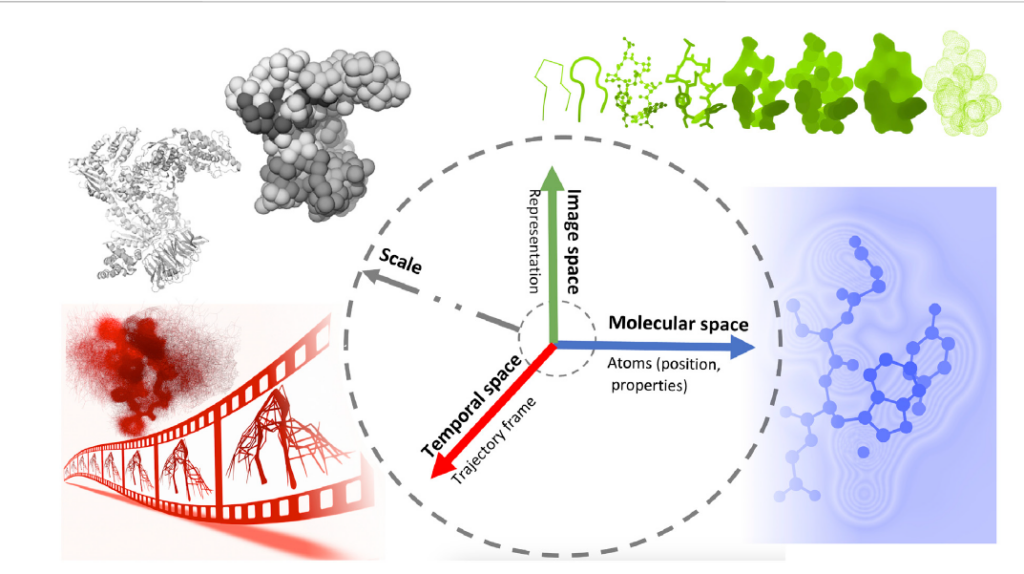
Advances in simulation, combined with technological developments in high-performance computing, have made it possible to produce physically accurate dynamic representations of complex biological systems involving millions to billions of atoms over increasingly longer simulation times. Analysis of these computational simulations is crucial, involving the interpretation of structural and dynamic data to gain insight into the underlying biological processes. However, this analysis is becoming increasingly challenging due to the complexity of the systems generated with a large number of individual runs, ranging from hundreds to thousands of trajectories. This massive increase in raw simulation data creates processing and visualization challenges. Effective visualization techniques play a crucial role in facilitating the analysis and interpretation of molecular dynamics simulations. In this paper, the authors focus on the techniques and tools that can be used to visualize molecular dynamics simulations, highlighting the few approaches specifically used for this purpose, discussing their advantages and limitations, and addressing the future challenges of molecular dynamics visualization.
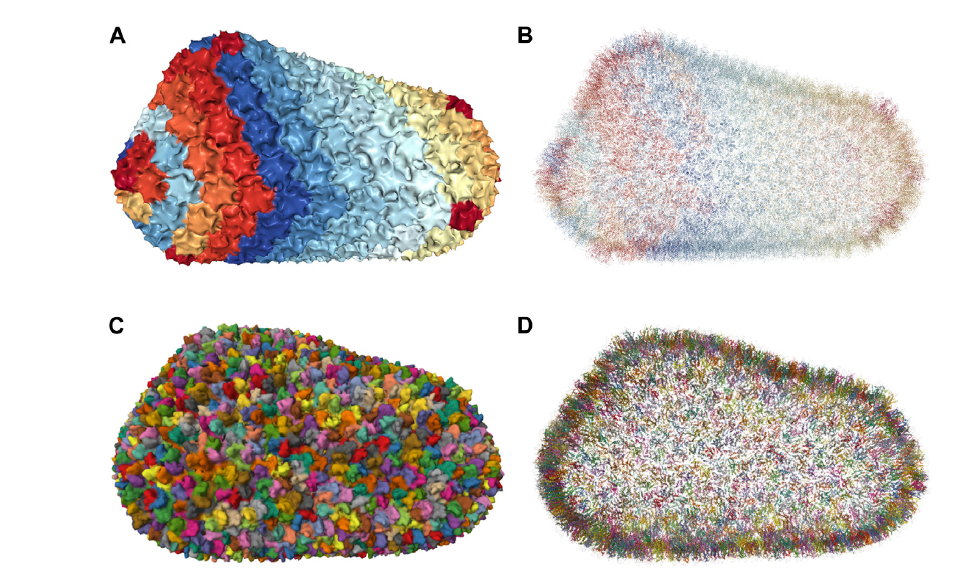
Cartoon representation of the HIV-1 capsid using Mol* Viewer (Rose and Hildebrand, 2015; Sehnal et al., 2021).




