Most membrane proteins are modified by the covalent addition of complex sugars through N- and O-glycosylation. Unlike proteins, glycans do not typically adopt specific secondary structures and remain highly mobile, potentially shielding large fractions of the protein surface. Their high degree of conformational freedom hinders the complete structural elucidation of glycoproteins. Computer simulations can be used to model glycosylated proteins but require hundreds of thousands of hours of computing time on supercomputers, limiting their routine use.
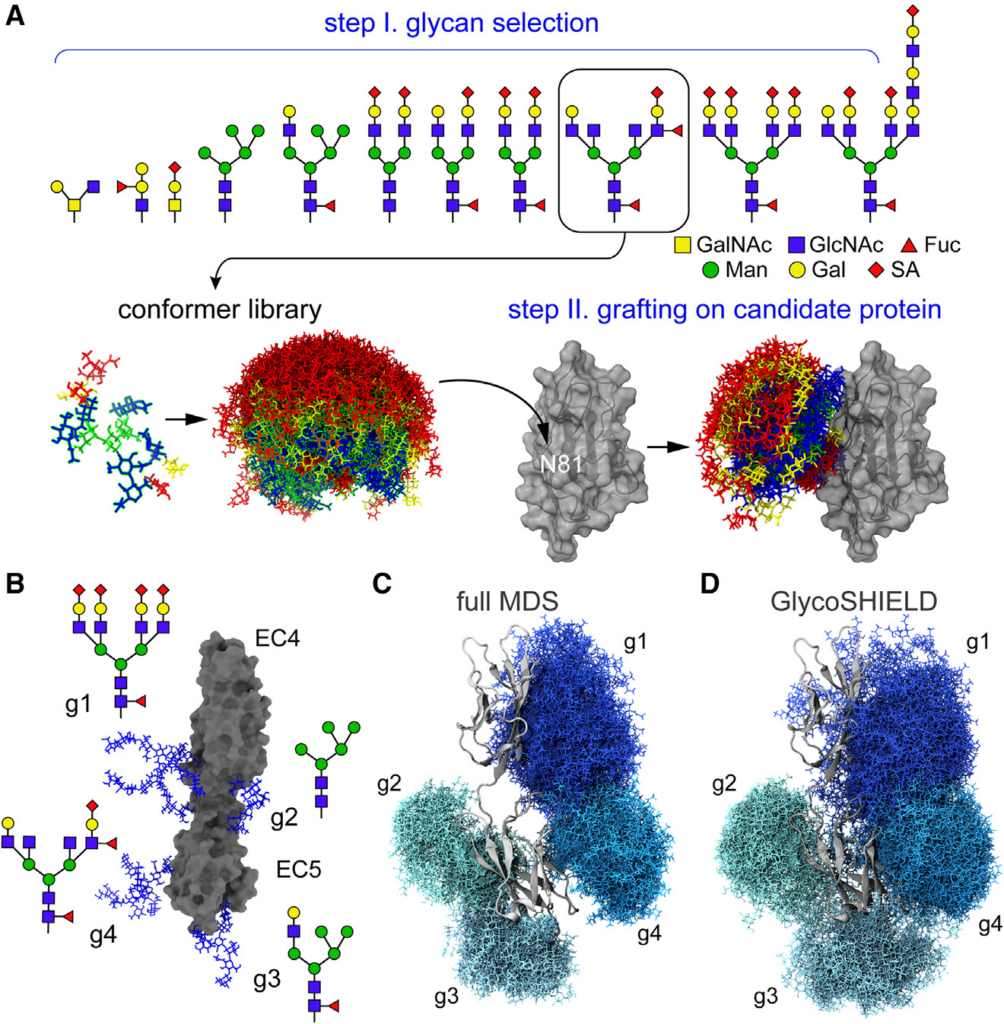
GlcNAc, N-acetylglucosamine; Fuc, fucose; Man, mannose; Gal, galactose; SA, sialic acid; see color code in inset). (B) Structure of N-cadherin EC4-EC5 model system (surface representation, gray) with four distinct N-glycans as indicated at each glycosylation site (sticks, blue, g1–g4). (C and D) Glycan conformers (sticks, shades of blue) generated by full MDS (C) or with GlycoSHIELD (D) after alignment on EC4-EC5 (cartoon, gray). Note the comparable morphology and span of the glycan shields obtained by the two approaches.
The authors describe GlycoSHIELD, a reductionist method that can be used on personal computers to realistic ensembles of glycan conformers onto static protein structures in a matter of minutes. Using molecular dynamics simulation, small-angle X-ray scattering, cryoelectron microscopy and mass spectrometry. The authors show that this open-access toolkit provides improved models of the structure of glycoproteins. Focusing on N-cadherin, the spike proteins of the human coronavirus and the gamma-aminobutyric acid receptor, the authors show that GlycoSHIELD can elucidate the influence of glycans on the conformation and activity of complex glycoproteins.




