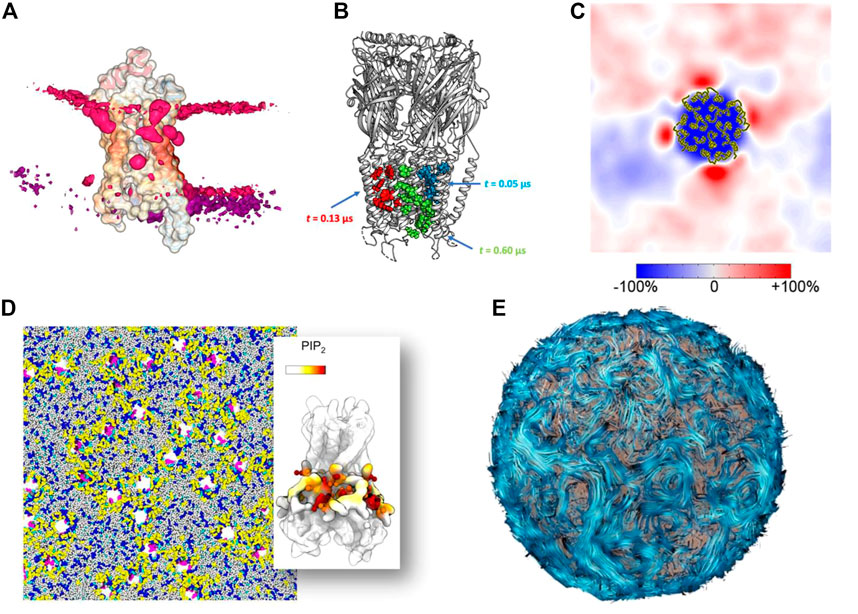
Understanding lipid dynamics and function, from the level of single, isolated molecules to large assemblies, is more than ever an intensive area of research. The interactions of lipids with other molecules, particularly membrane proteins, are now extensively studied. With advances in the development of force fields for molecular dynamics simulations (MD) and increases in computational resources, the creation of realistic and complex membrane systems is now common. In this perspective, we will review four decades of the history of molecular dynamics simulations applied to membranes and lipids through the prism of molecular graphics.