Carbohydrate molecules present in more than 14,000 Protein Data Bank (PDB) structures have recently been reviewed and remediated to conform to a new standardized format. This machine-readable data representation for carbohydrates occurring in the PDB structures and the corresponding reference data improve the findability, accessibility, interoperability and reusability of structural information pertaining to these molecules.
The PDB Exchange MacroMolecular Crystallographic Information File data dictionary now supports:
(i) standardized atom nomenclature that conforms to International Union of Pure and Applied Chemistry-International Union of Biochemistry and Molecular Biology (IUPAC-IUBMB) recommendations for carbohydrates
(ii) uniform representation of branched entities for oligosaccharides
(iii) commonly used linear descriptors of carbohydrates developed by the glycoscience community
(iv) annotation of glycosylation sites in proteins.
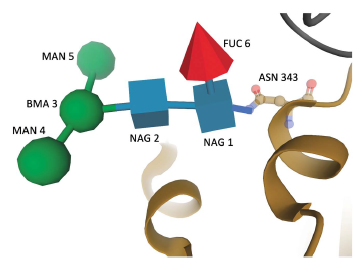
For the first time, carbohydrates in PDB structures are consistently represented as collections of standardized monosaccharides, which precisely describe oligosaccharide structures and enable improved carbohydrate visualization, structure validation, robust quantitative and qualitative analyses, search for dendritic structures and classification. The uniform representation of carbohydrate molecules in the PDB described herein will facilitate broader usage of the resource by the glycoscience community and researchers studying glycoproteins.




