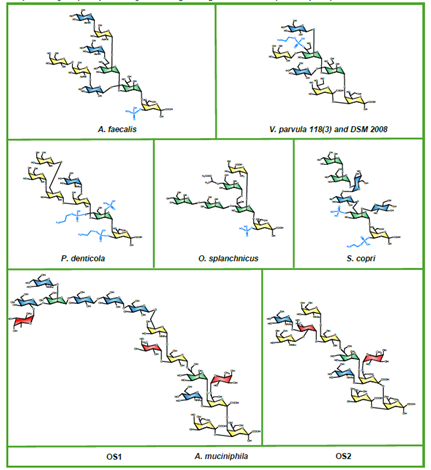
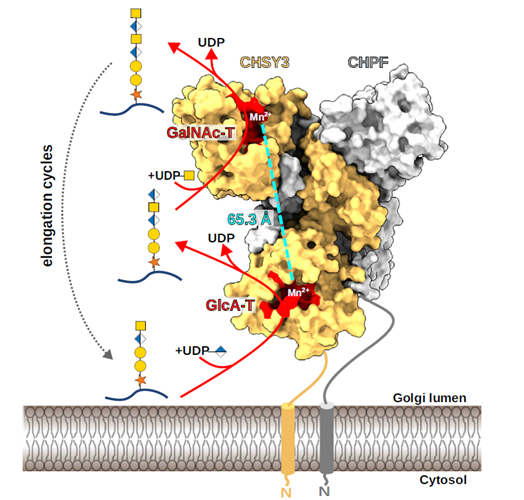
The transformative artificial intelligence (AI) tool called AlphaFold, has predicted the 3 dimensional structures of nearly the entire human proteome (98.5% of human proteins). The resulting dataset covers 58% of residues with a confident prediction, of which a subset (36% of all residues) have very high confidence. Furthermore, the tool has predicted almost complete proteomes for various other organisms, ranging from mice and maize (corn) to the malaria parasite.
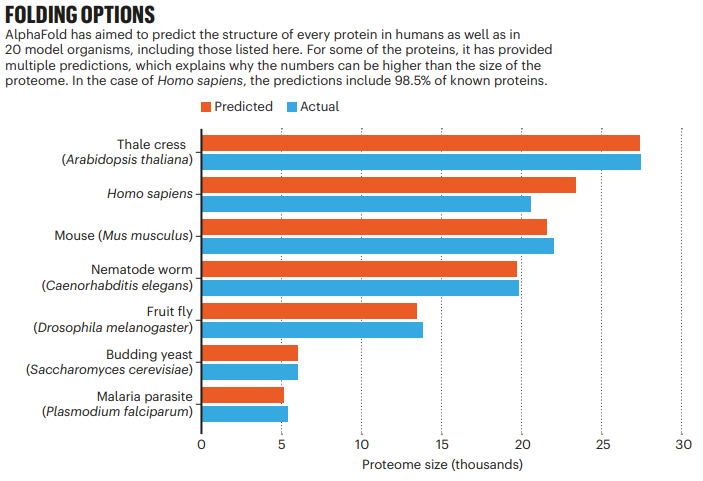
The predicted 3 dimensional structures are freely available to the community via a public database hosted by the European Bioinformatics Institute at https://alphafold.ebi.ac.uk/.