The new Glyco3D portal (glyco3D@cermav.cnrs.fr) brings together databases and applications of interest to structural glycosciences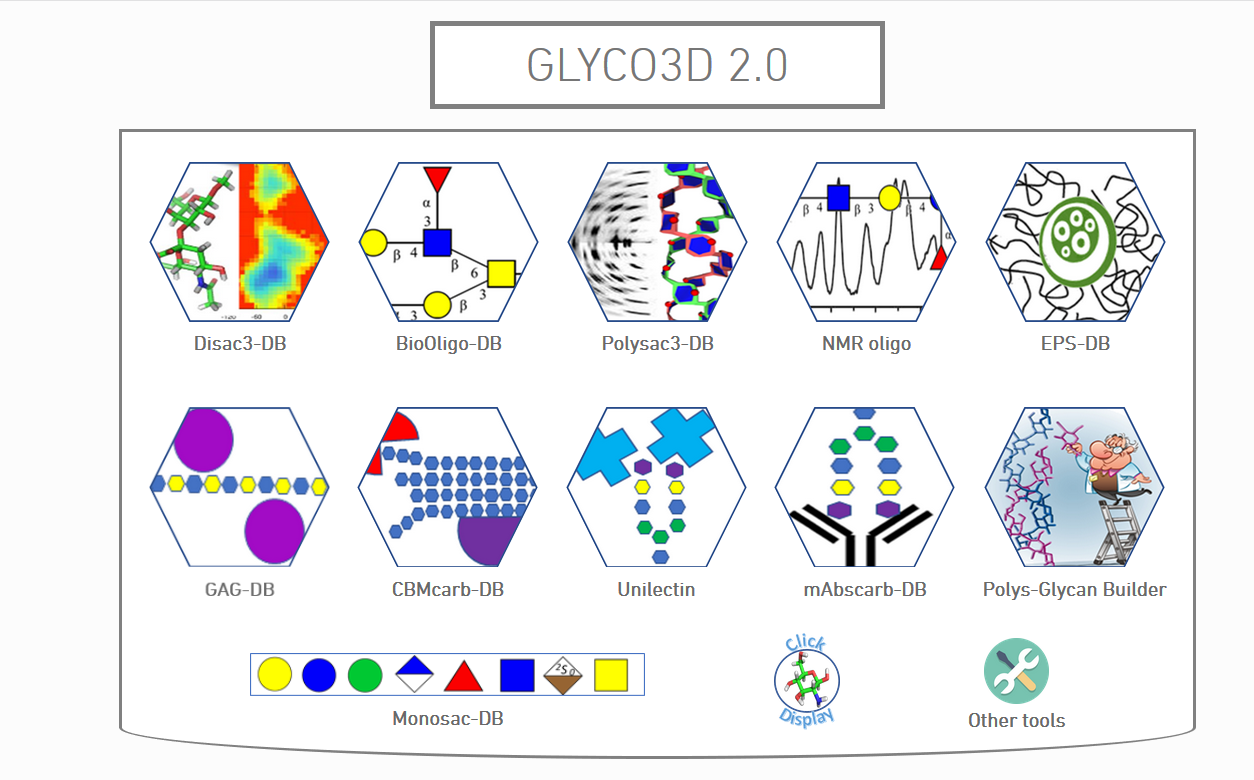
Disac3-DB: 160 disaccharides, 3D Structures, xyz, Potential Energy Surfaces, SNFG representations
BioOligo-DB : 314 oligosaccharide, 3D structures, xyz, Taxonomy, SNFG representations
Polysac3-DB : Polysaccharides, 3D Structures, xyz, X-ray, e- diffractograms, taxonomy, (curated)
NMR Oligo : NMR data, 150 oligosaccharides, 1H, 13C, COSY, TOCSY, HMQC & HMBC , Taxonomy, SNFG representations
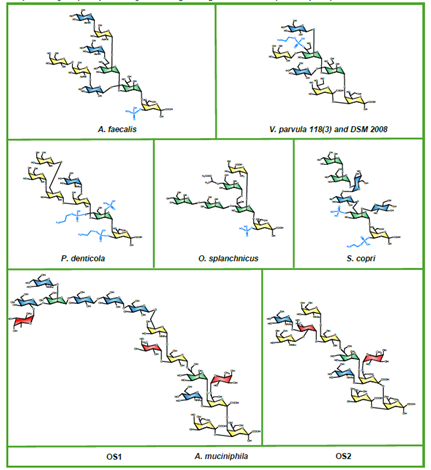
EPS-DB : 140 bacterial Polysaccharides; 3D structures, xyz, functional properties, genetic data, taxonomy (curated)
GAG-DB : 123 3D Xay of GAG binding proteins crystal complexes, xyz, Taxonomy, Bibliography (curated)
CBMcar-DB : 353 3D Xay of Carbohydrate Binding Modules crystal complexes, xyz, Taxonomy, Bibliography (curated)
Unilectin :2332 3D Xray lectins (1493 interacting glycan), xyz, Taxonomy, Bibliography (curated)
mAbscarb-DB : 3D XR mAb interacting with glycan, xyz, SNFG representation
Polys-Glycan Builder: 3D structure generation of poly- and complex oligosaccharides from MM2-precalculated glycosidic linkage torsions and energy minimization
Monosac-DB : 250 Monosaccharides, 3D structures, xyz, SNFG representations
Click & Diplay : Easy display by LiteMol from pdb
Other Tools
glyco3D@cermav.cnrs.fr