The ongoing COVID-19 pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Antibody development efforts mainly revolve around the extensively glycosylated SARS-CoV-2 spike (S) protein, which mediates host cell entry by binding to the angiotensin-converting enzyme 2 (ACE2). Like many other viral fusion proteins, the SARS-CoV-2 spike utilizes a glycan shield to thwart the host immune response. The authors built a full-length model of the glycosylated SARS-CoV-2 S protein, both in the open and closed states, augmenting the available structural and biological data. Multiple microsecond-long, all-atom molecular dynamics simulations were used to provide an atomistic perspective on glycans’ roles and o the protein structure and dynamics. The results reveal N-glycans’ essential structural role at sites N165 and N234 in modulating the conformational dynamics of the spike’s receptor-binding domain (RBD), which is responsible for ACE2 recognition.
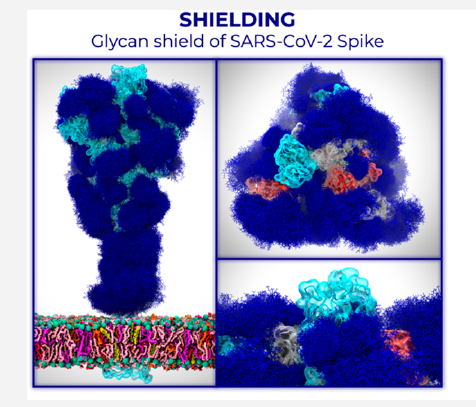
Bilayer interferometry experiments corroborate this finding. They show that deletion of these glycans through N165A and N234A mutations significantly reduces binding to ACE2 due to the RBD conformational shift toward the “down” state. Additionally, end-to-end accessibility analyses outline a complete overview of the glycan shield’s vulnerabilities of the SARS-CoV-2 S protein, which may be exploited in the therapeutic efforts targeting this molecular machine. Overall, this work presents hitherto unseen functional and structural insights into the SARS-CoV-2 S protein and its glycan coat, providing a strategy to control the conformational plasticity of the RBD that could be harnessed for vaccine development.