The heterogeneity, mobility and complexity of glycans in glycoproteins have been, and currently remain, significant challenges in structural biology.
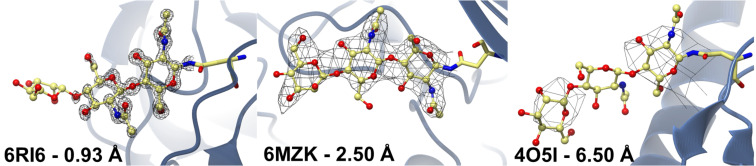
These aspects present unique problems to the two most prolific techniques: X-ray crystallography and cryo-electron microscopy. At the same time, advances in mass spectrometry have made it possible to get deeper insights on precisely the information that is most difficult to recover by structure solution methods: the full-length glycan composition, including linkage details for the glycosidic bonds. The developments have given rise to glycomics. Thankfully, several large scale glycomics initiatives have stored results in publicly available databases, some of which can be accessed through API interfaces. In the present work, we will describe how the Privateer carbohydrate structure validation software has been extended to harness results from glycomics projects, and its use to greatly improve the validation of 3D glycoprotein structures.




