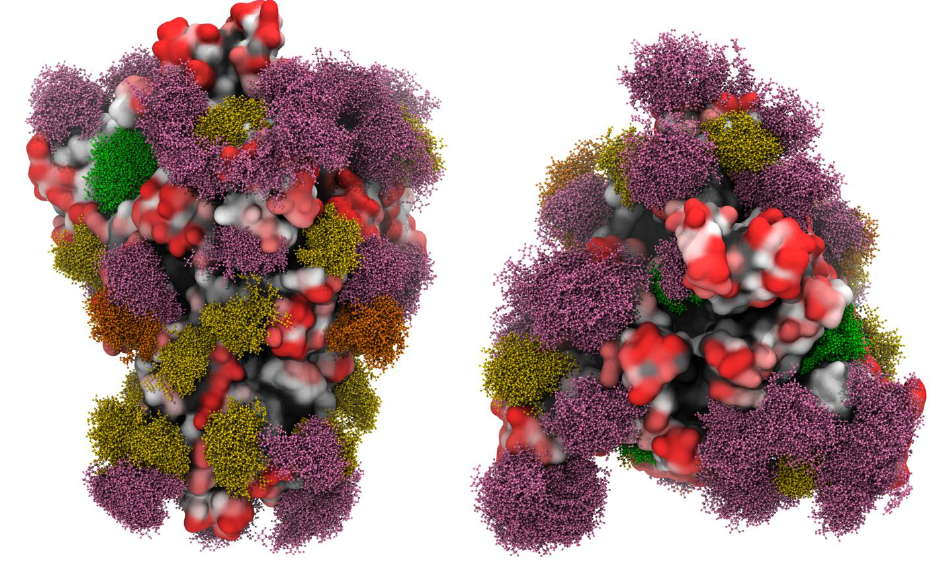
The determination of the three-dimensional structure fo the spike (S) protein SARS-CoV-2, along with reported glycomics data for the protein produced in HEK293 cells indicated an extensive level of glycosylation but did not provide any structural information as regards to the role played by these glycans.
The authors of the article report the structures for glycoforms that represent those present in the nascent glycoproteins (prior to enzymatic modifications in the Golgi and ER), as well as those that are commonly observed on antigens present in other viruses.
These models were subjected to MD simulation to take into account protein and glycan plasticity and to determine the extent to which glycan microheterogeneity impacts antigenicity. Another feature of the work deals with the identification of the peptides in the S protein that are likely to be presented in human leukocyte antigen (HLA) complexes. A follow-up discussion addresses the role of S protein glycosylation in potentially modulating the adaptive immune response to the SARS-CoV-2 virus or to a related vaccine.