It is fully recognized that no symbol system will ever convey a full appreciation of the three-dimensional structures of glycans, essential information for the understanding of how glycans and proteins interact (A. Varki et al., 2008). This is clearly illustrated by the example given in Fig. 11 which shows the many monosaccharide units that can satisfy the stereochemical requirements resulting from the spatial arrangements of 3 contiguous hydroxyl groups, for recognition by the binding site of the PA-IIL lectin from Pseudomonas aeruginosa.
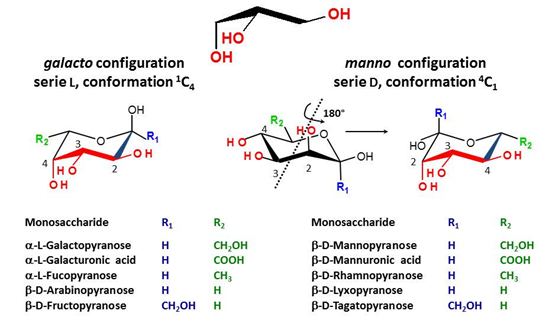
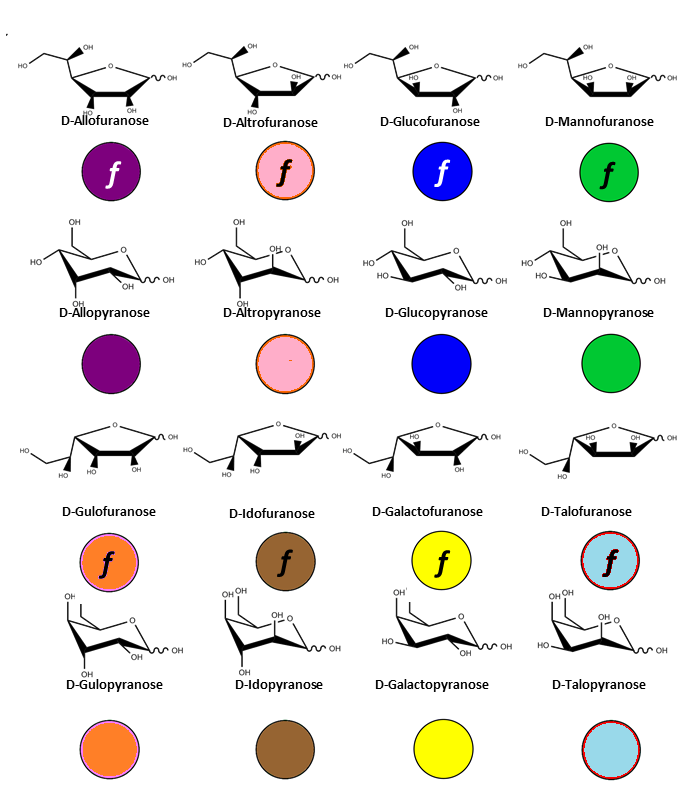
There is indeed a need to link the symbol notation to a graphical representation that maintains the stereochemical feature of each monosaccharide. The results of such an exercise have been reported (O. Berger et al., 2008). Typical representations of symbol notations and graphical representations are given for cyclic furanose and pyranose forms :
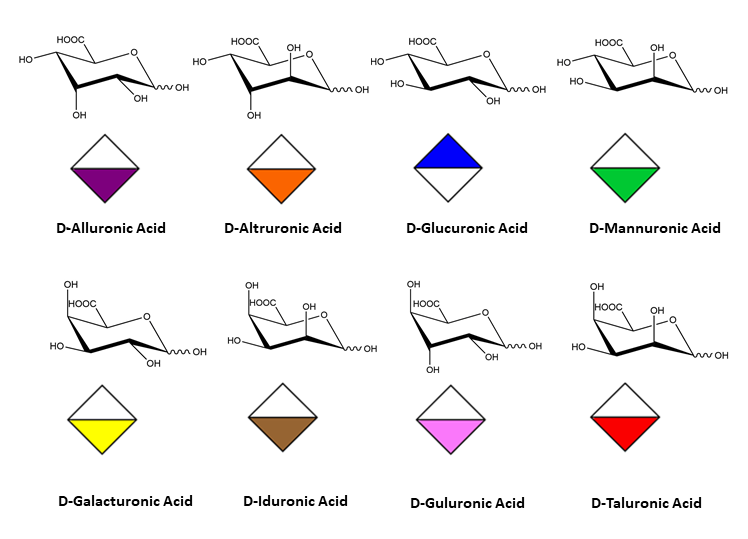
cyclic forms of hexopyranuronic acids
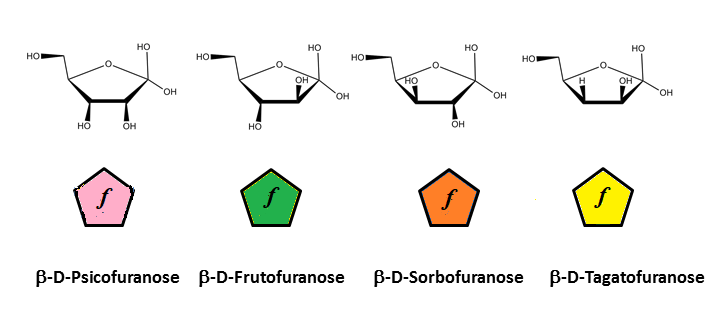
cyclic forms of 2-ketoses
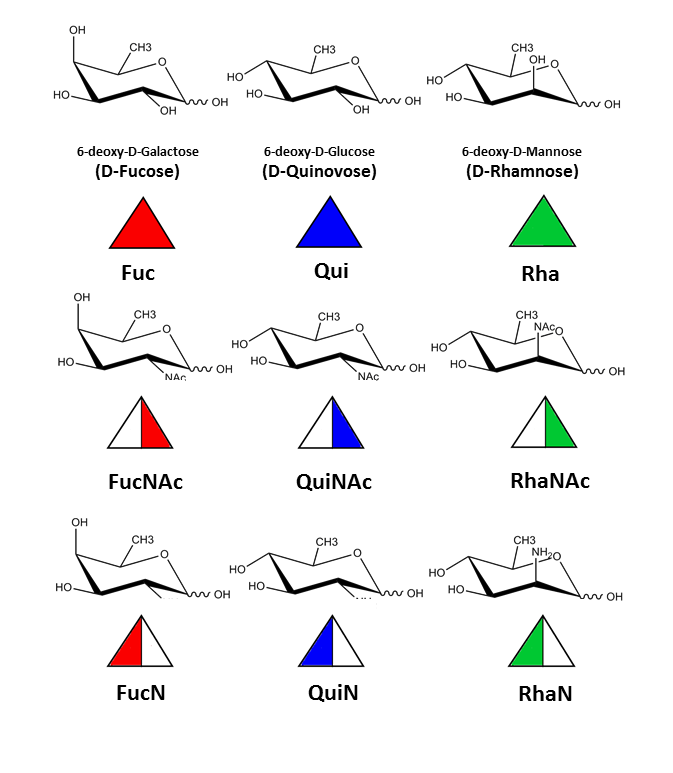
three-representatives of 6-deoxy sugars, with NAc and N at the position 2
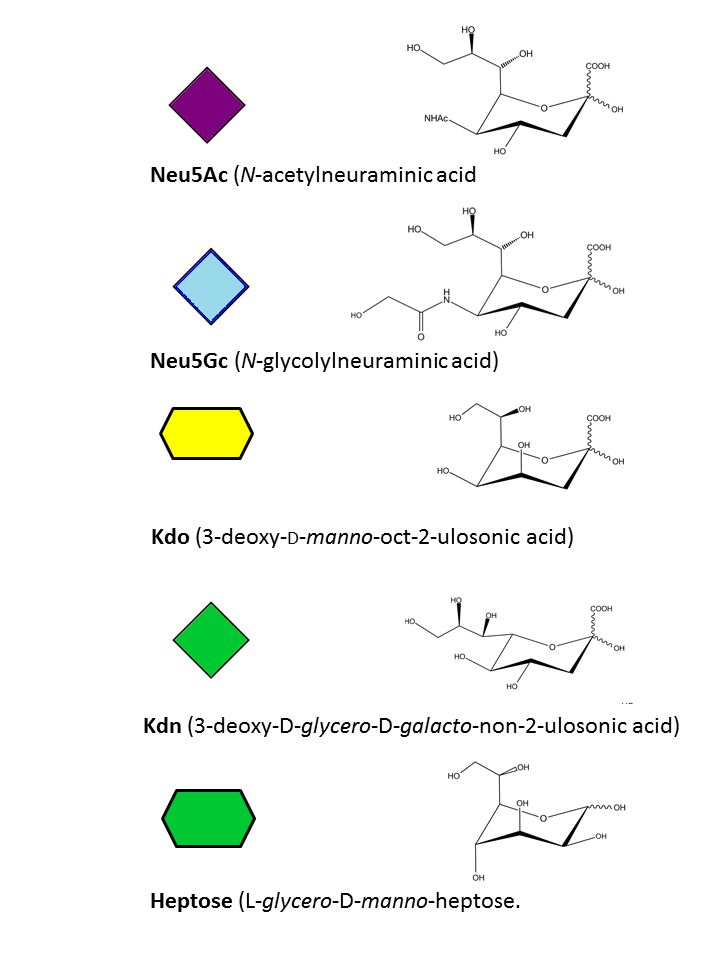
Neu5Ac, Neu5Gc, Kdo, Kdn, Heptose
Encoding and Representation
The most popular ways of encoding and representing glycans are:
- – Linear notation (e.g. IUPAC)
- – Symbolic / diagrammatic notation (e.g. Oxford)
- – The notation proposed by the Consortium for Functional Glycomics www.functionalglycomics.org.
It is nevertheless possible to concatenate the symbolic representation of monosaccharides with a limited number of structural descriptors to achieve a fairly exhaustive nomenclature which can be further used in constructing three dimensional structures of glycans. This can be achieved using the following set :
Residue Letter Name: Rib, Ara, Xyl, Lyx, All, Alt, Glc, Man, Gul, Ido, Gal, Tal,…. and abbreviated trivial name.
[O-ester and ethers]: (when present) are shown attached to the symbol with a number, e.g.
- 6Ac for 6-O-acetyl group
- 3S for 3-O-sulfate group
- 6P for 6-O-phosphate group
- 6Me for 6-O-Methyl group
- 36Anh for 3,6-anhydro
- Pyr for pyruvate group
Absolute Configuration: D or L
The D-configuration for monosaccharide and the L configuration for Fucose and Idose are implicit and does not appear in the symbol. Otherwise the L configuration, is indicated in the symbol, as in the case of Arabinose or L-Galactose.
For those occurring in the furanose form, a letter N or S is inserted in the symbol, indicating the northern (N) or Southern (S) conformation of the five membered ring.
Anomeric configuration: The nature of the glycosidic configuration (α or β) is explicitly set within the symbol.
Ring conformation
All pyranoses in the D-configuration are assumed to have 4C1 chair conformation; those in the L configuration are assumed to have 1C4 chair conformation. Otherwise, the ring conformation is indicated in the symbol, as 2S0 in the case of α-L-Idopyranose.
N or S indicates the conformation of the five membered rings on the conformational wheel.
While maintaining the spirit of using the symbolic representation for monosaccharides (and towards glycans) this set of rules provides the necessary extension to the construction of three-dimensional structures, allowing encoding for computational manipulation whilst maintaining IUPAC nomenclature.
