The multiple monosaccharide building blocks can be linked to various regio-chemistries and stereo-chemistries, and the resulting glycan be assembled on protein or lipid scaffolds. The diversity of glycoconjugate structures comprises an “information-rich” system capable of participating in a wide range of biological functions. There comes the question of representing and encoding the many monosaccharides in ways that meet the practices of several scientific communities and are compatible with the requirement of bioinformatics.
Like nucleic acids and proteins, it is far more efficient to encode glycan using a residue-based approach. However, as compared to nucleic acids or proteins, there are a far greater number of residues arising, due to the frequent modifications occurring on the parent monosaccharides. Also, since glycans are frequently found to have branched structures, most of them are tree-like molecules, unlike nucleic acids and proteins. The pre-requisite for a residue-based encoding format is a controlled vocabulary of residue names. For practical reasons, it makes sense to restrict the number of residues to as low a number as possible. Yet the lack of clear rules to subscribe atoms of a molecule to one particular monosaccharide and not of a substituent, pose the main hurdle in encoding monosaccharide names. The variety in nomenclature and structural representation of glycans makes it difficult to decide the most appropriate form of illustrating the approach of the scientific investigation. The choice of notation may be based on whether the study is focused on the chemistry or on the biology side of glycoscience. Moreover, the information content of each representation may vary or highlight a particular aspect as compared to others. While representing a complex glycan structure, chemists could favor a representation that includes information about the anomeric carbon, the chirality, the monosaccharides present along with the glycosidic linkages connecting them. For others, it could be more appropriate to visualize the monosaccharides present, and hence a symbolic/diagrammatic representation would be favored.
The Symbol and Text Nomenclature for Representation of Glycan Structures.
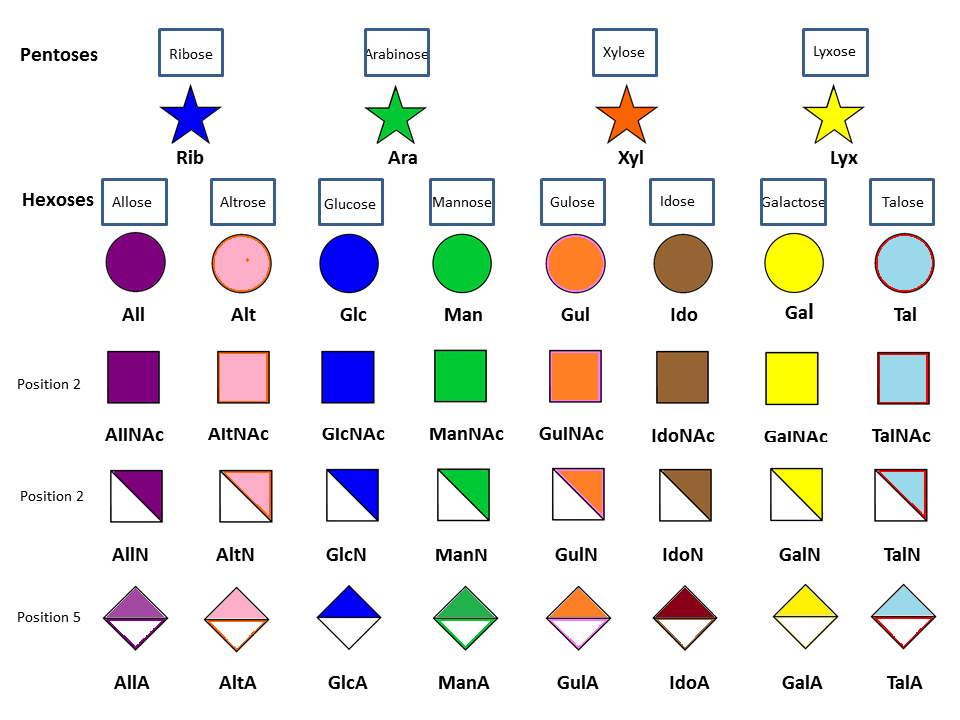
The representation of monosaccharides as symbols is a result of a concerted effort under the umbrella of the Consortium for Functional Glycomics (CFG). (www.functionalglycomics.org). After evaluating widely used symbol nomenclatures, an ad hoc nomenclature committee selected a version originally put forth by Stuart Kornfeld and later adapted by the editors of the textbook ’Essentials of Glycobiology’. Further considerations were concerned with the adequacy for the annotation of mass spectra, as well as achieving common ground with as many other interested groups as possible. It resulted in the following set of rules :
- Avoid same shape/color but a different orientation to represent different sugars since annotations of mass spectra are hard to show as horizontal cartoons.
- Choice of symbols should be logical and simple to remember. Each monosaccharide type (e.g. Hexose) should have the same shape, and isomers are to be differentiated by color /black/white/shading.
- Use same shading/ color for different monosaccharides of same stereochemical designation e.g., Gal, GalNAc, GalA should all be the same shading/color.
- To minimize variations, sialic acids and uronic acids are the same shapes. Only major uronic and sialic acid types are represented.
- While color is useful, the system should also function with black and white, and colored representations should survive black-and-white printing or photocopying
- Positions of linkage origin are assumed to be common ones unless indicated. Anomeric notation and destination linkage indicated without spacing/dashes.
- Modifications of monosaccharides (e.g., sulfation, O-acetylation) indicated by attached small letters, with numbers indicating linkage positions, if known.
The symbolic representation dealing essentially with mammalian glycans has gained wide acceptance and is being used to describe glycan composition and biosynthesis as illustrated for N- and O-linked glycans.
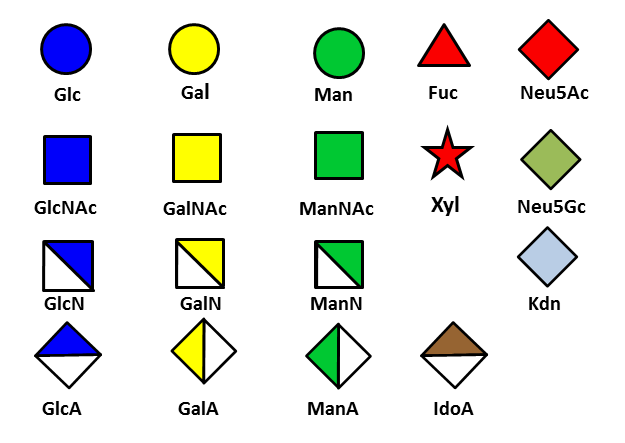
The symbolic representation dealing essentially with mammalian glycans has gained wide acceptance and is being used to describe glycan composition and biosynthesis as illustrated for N- and O-linked glycans.
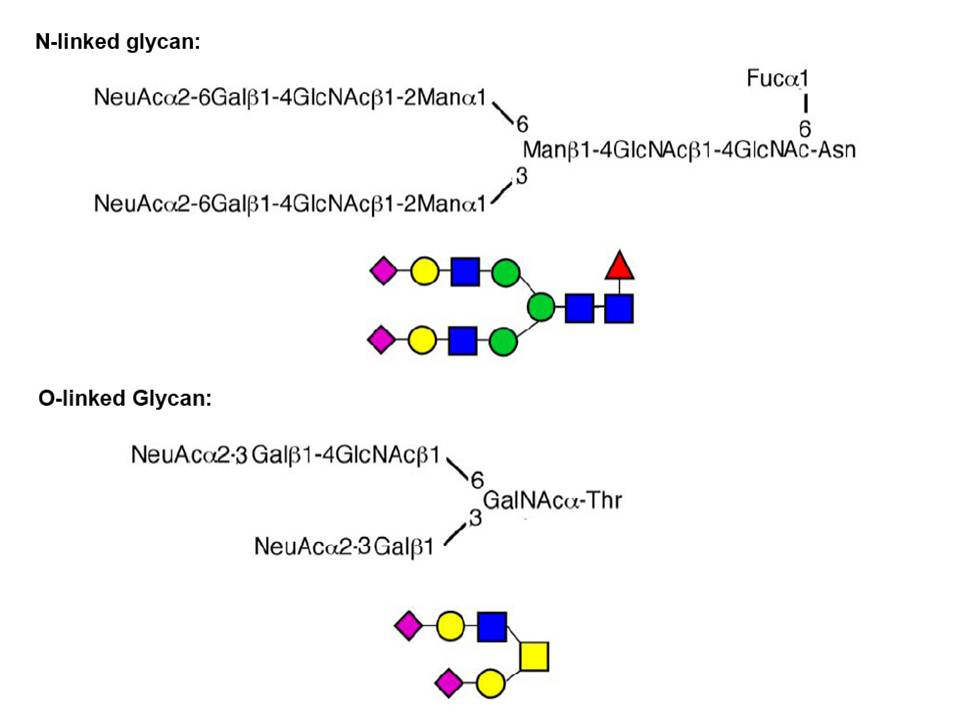
The preliminary set of symbol notation for carbohydrates dealt with a limited number of monosaccharide units, which indeed provides a satisfactory coverage to describe the mammalian oligosaccharides which have been characterized so far.
The Extension of the Symbol Notation to More Complex Glycans
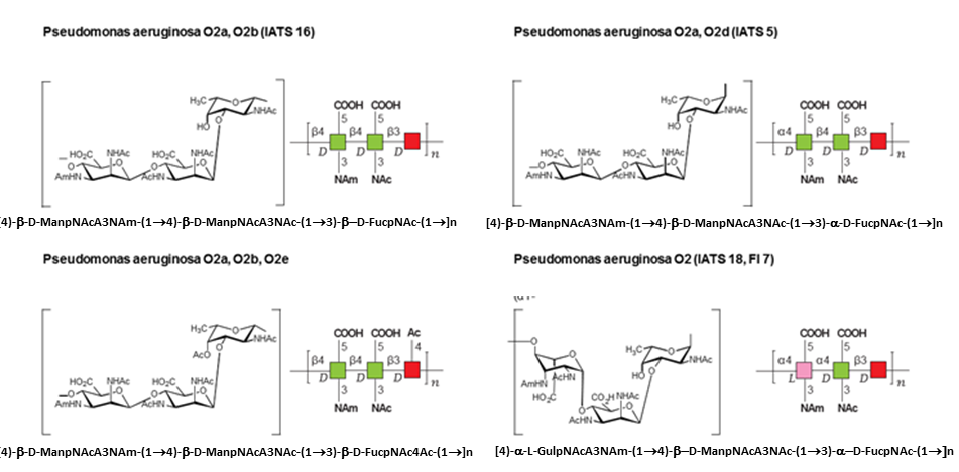
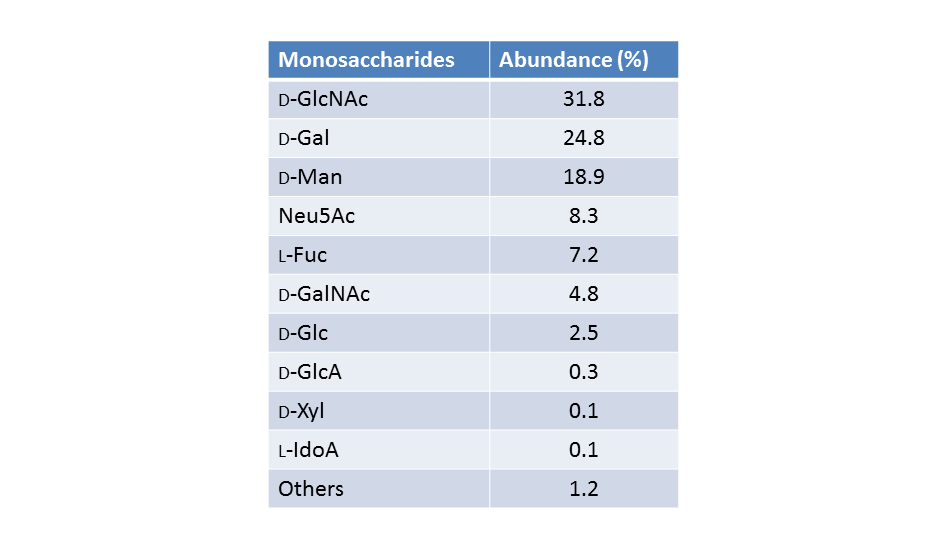
The symbol notation has been extended to describe the structure of pathogen polysaccharides (O. Berger et al., 2008) with the objective of providing a quick and easy way to visually distinguish between polysaccharides, for understanding cross-reactivity of sera on pathogen glycan arrays. Those glycans that are encountered in pathogens are more diverse and contain greater structural variability than glycans found in mammals. There is a need to identify monosaccharides (by shape and color) and with the connectivity and substituent differences between polysaccharides. An extra level of complexity is found, occurring from the fact that pathogens utilize a greater diversity of monosaccharides with multiple modifications, which can occur both in the D- and L-configurations, and may be found in both pyranose and furanose forms.