While maintaining the spirit of using the symbolic representation for monosaccharides (and towards glycans) this set of rules provides the necessary extension to the construction of three-dimensional structures, allowing encoding for computational manipulation whilst maintaining IUPAC nomenclature.
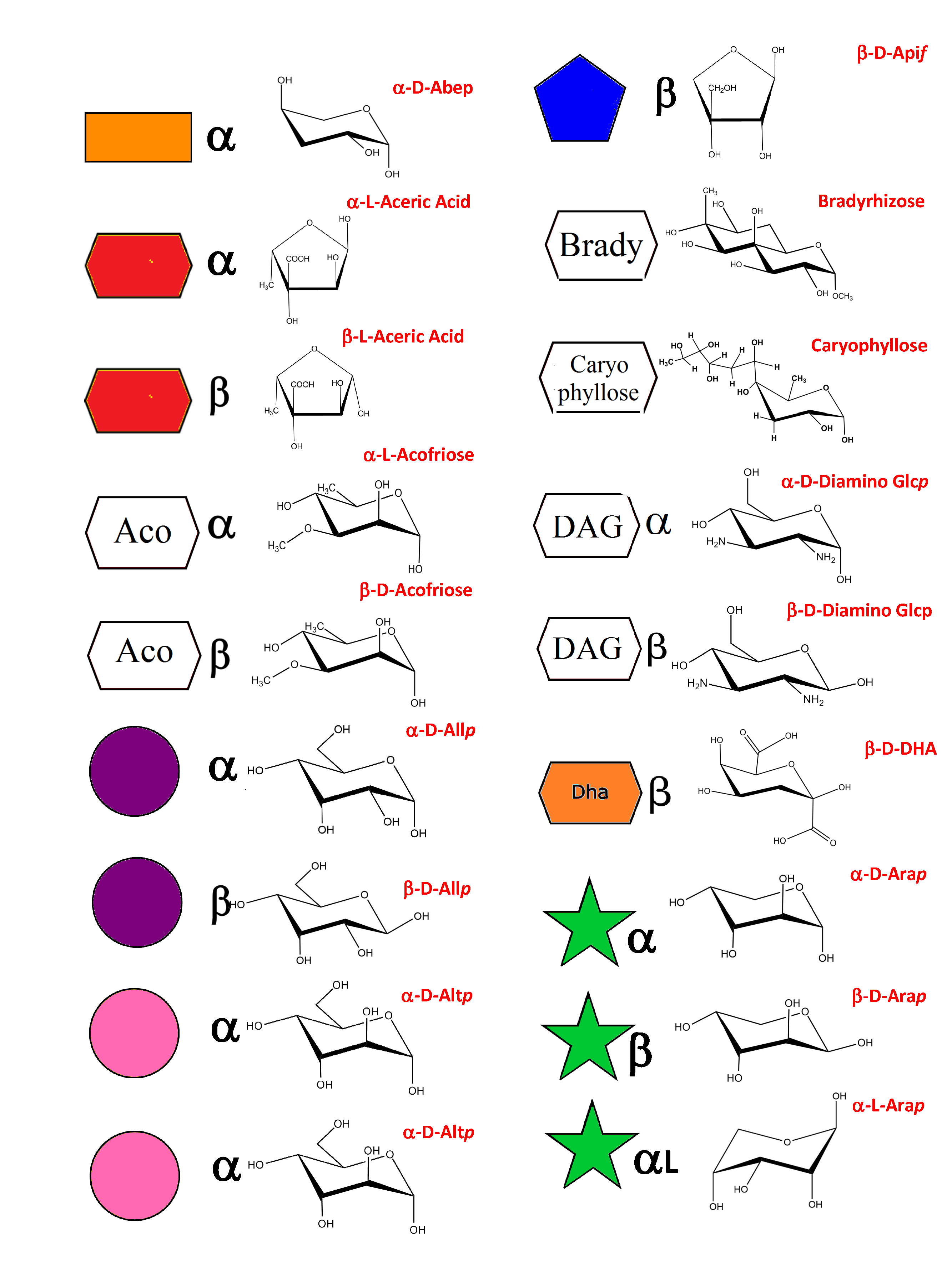
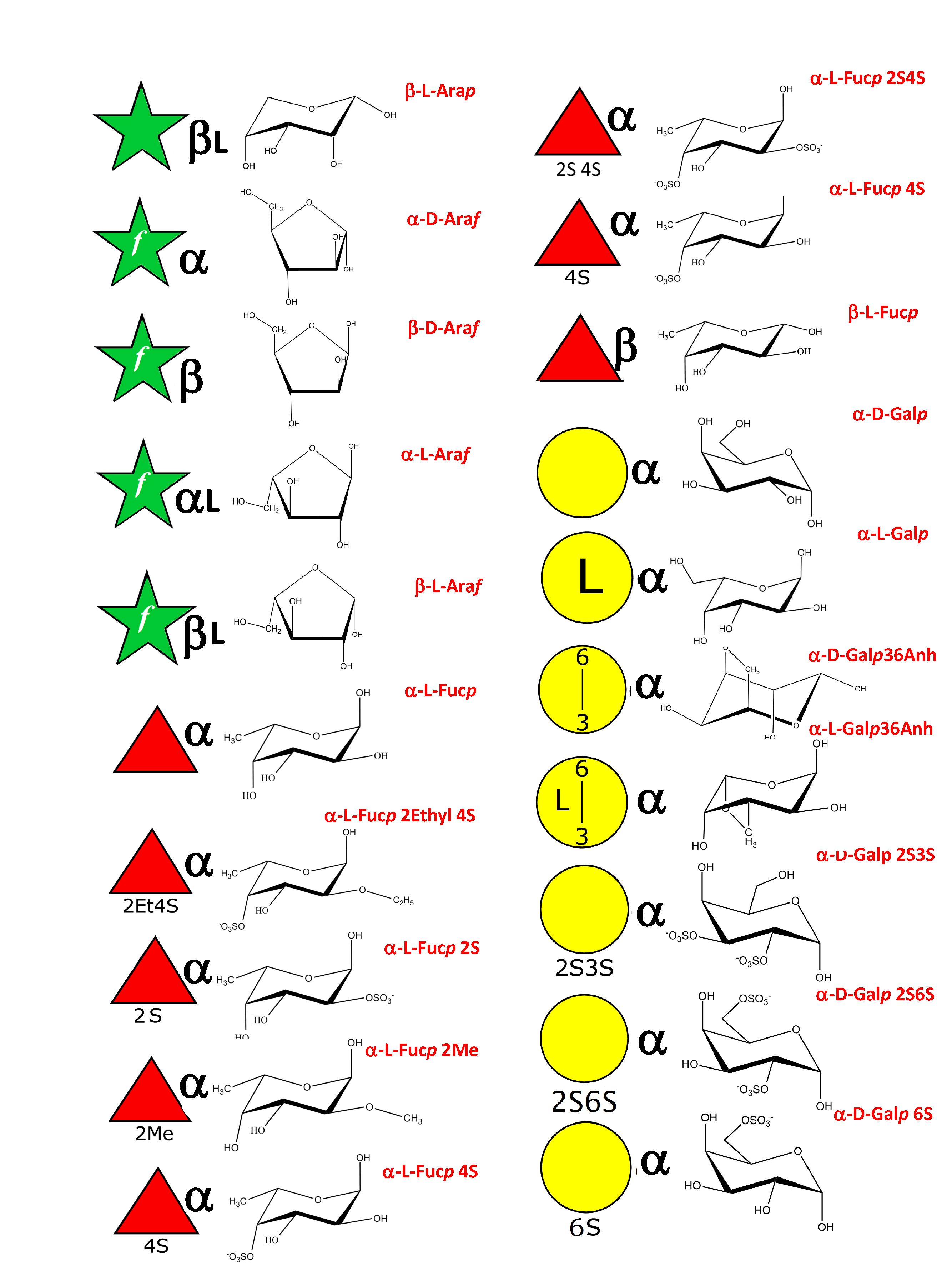
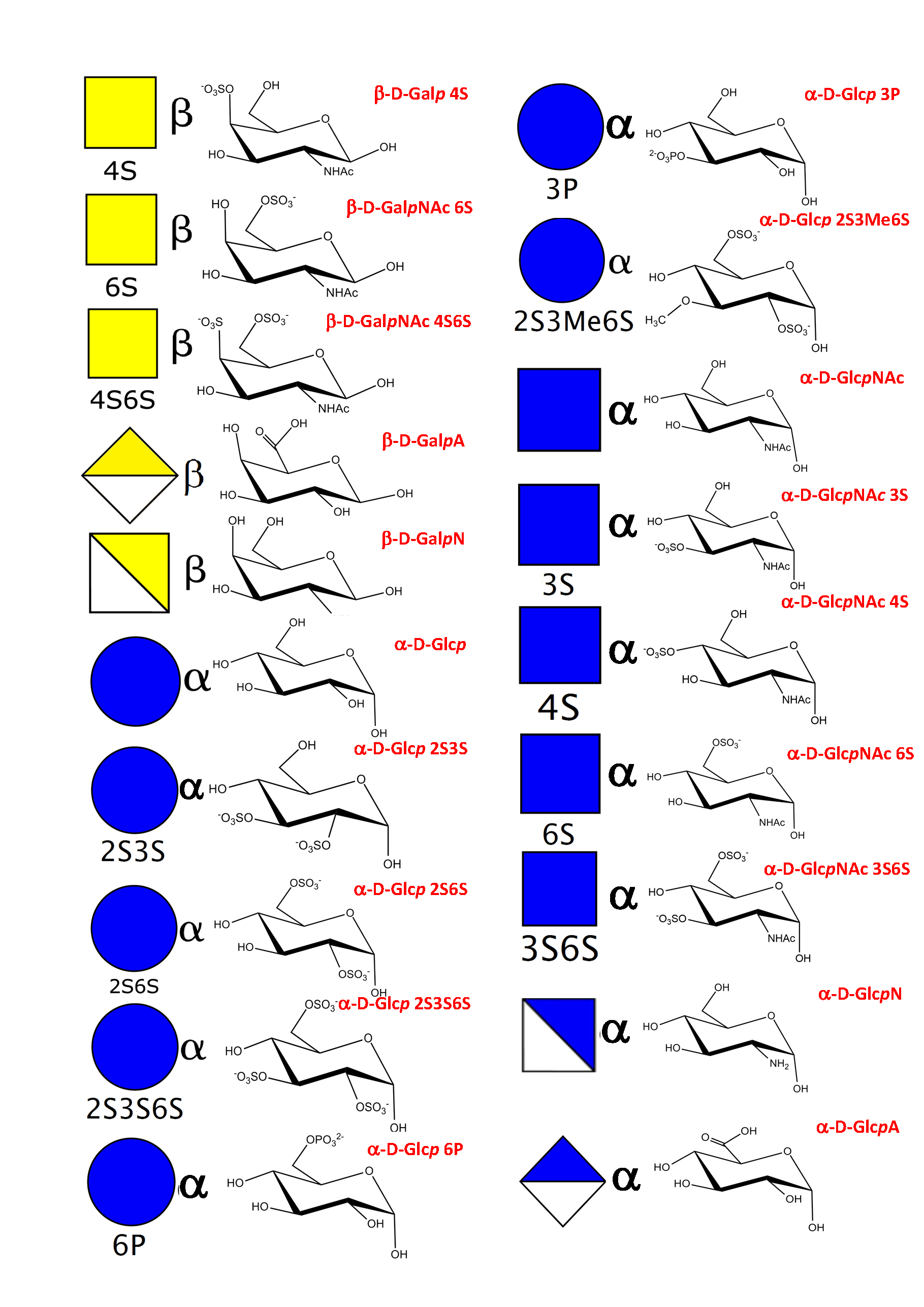
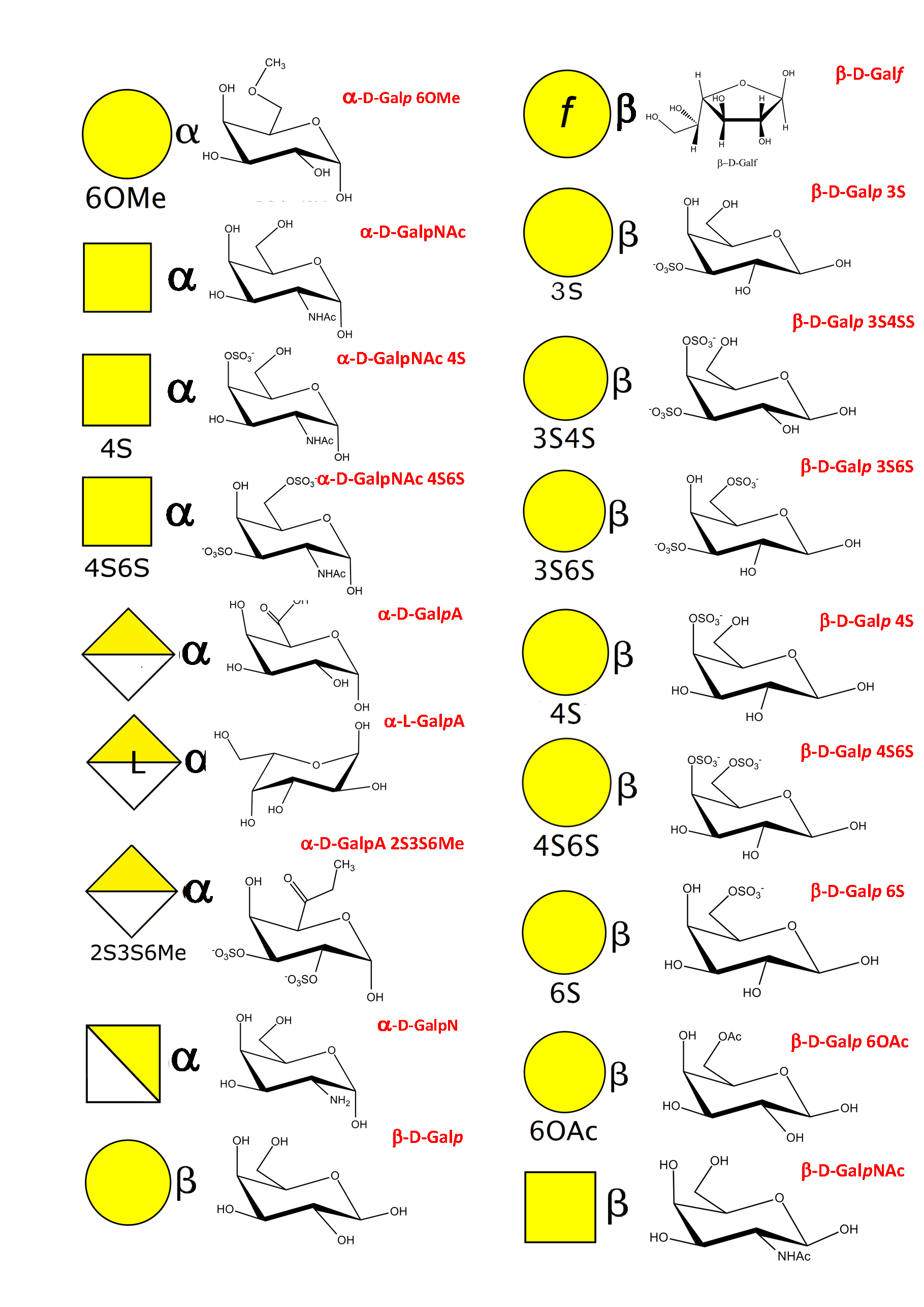
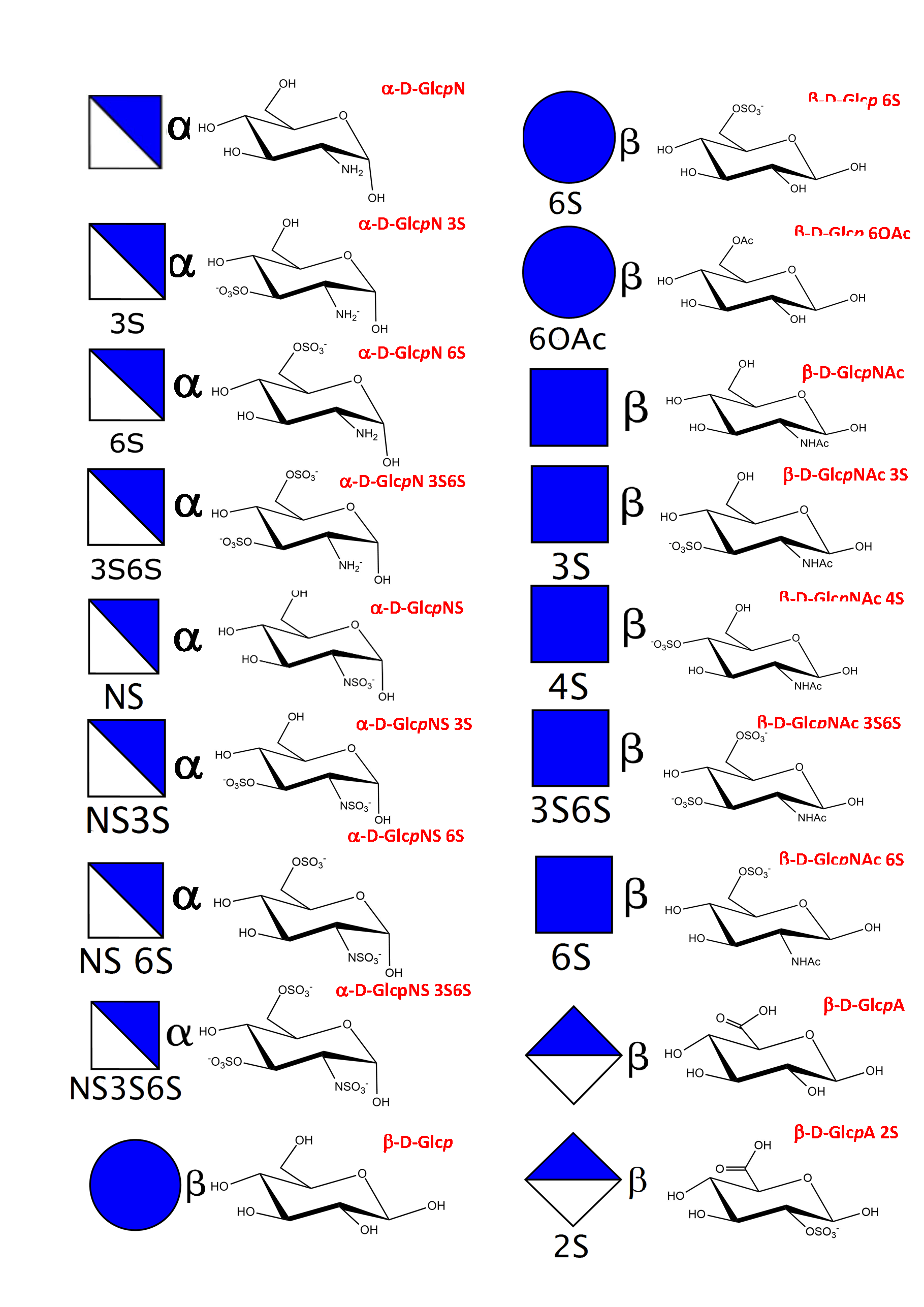
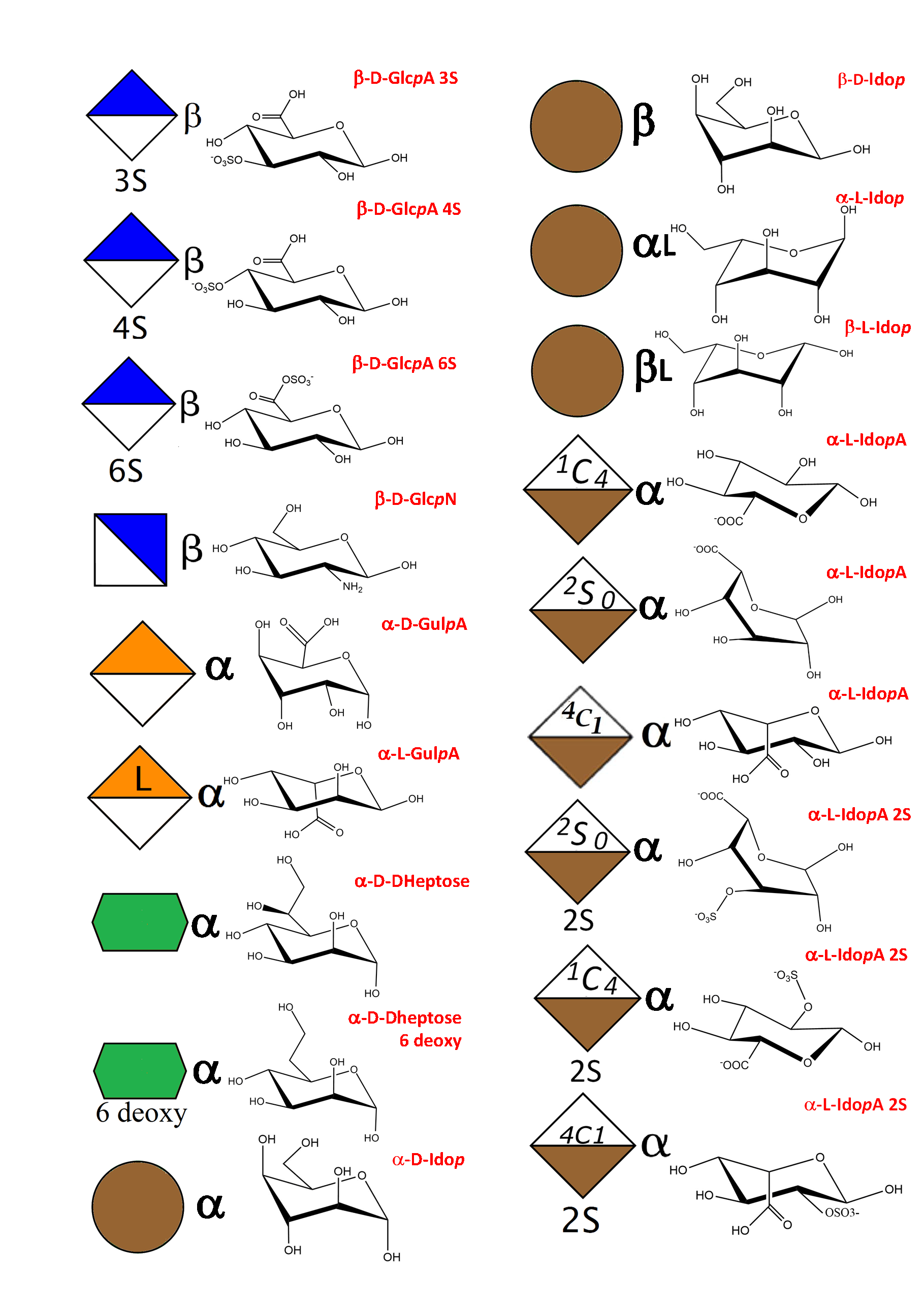
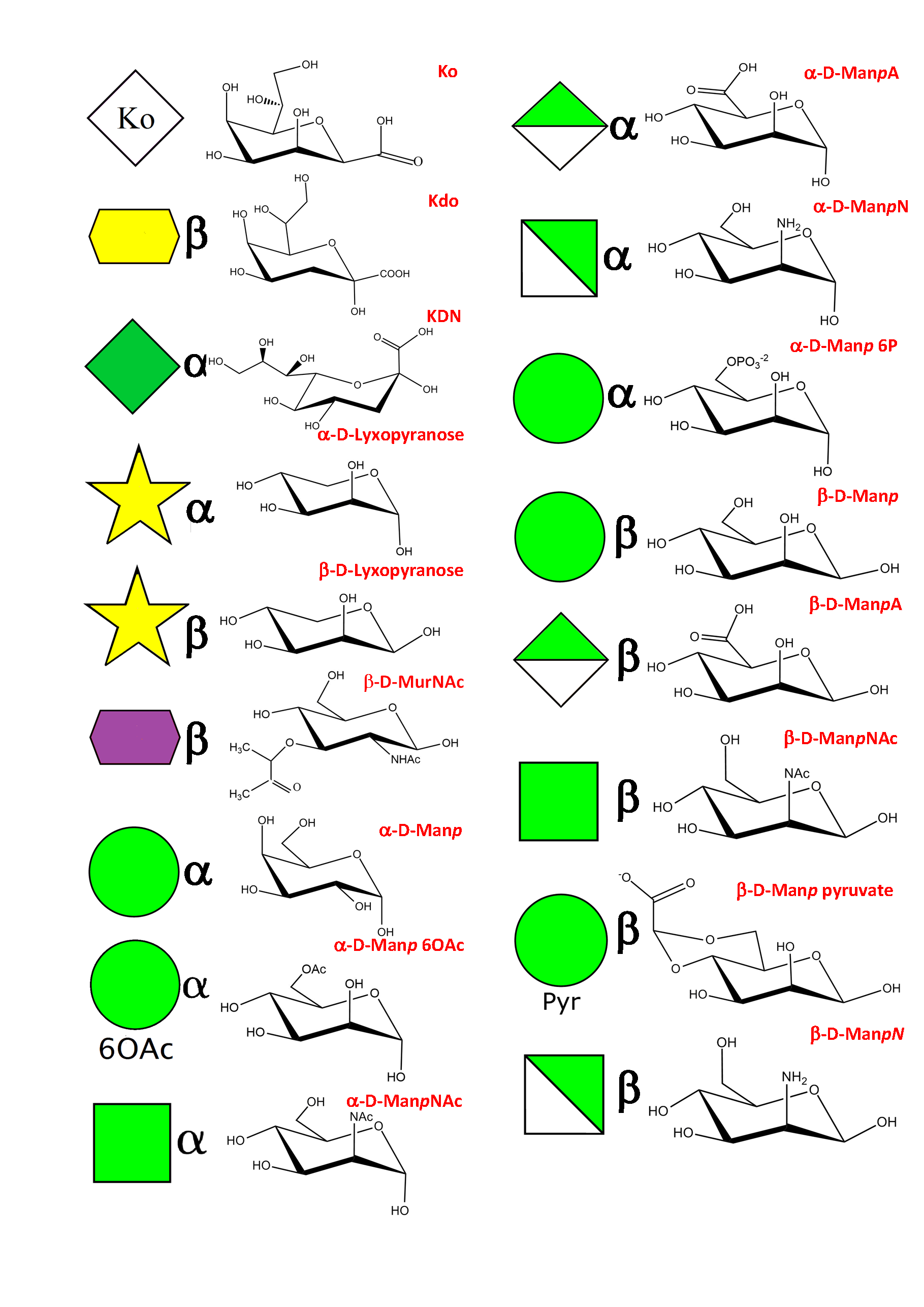
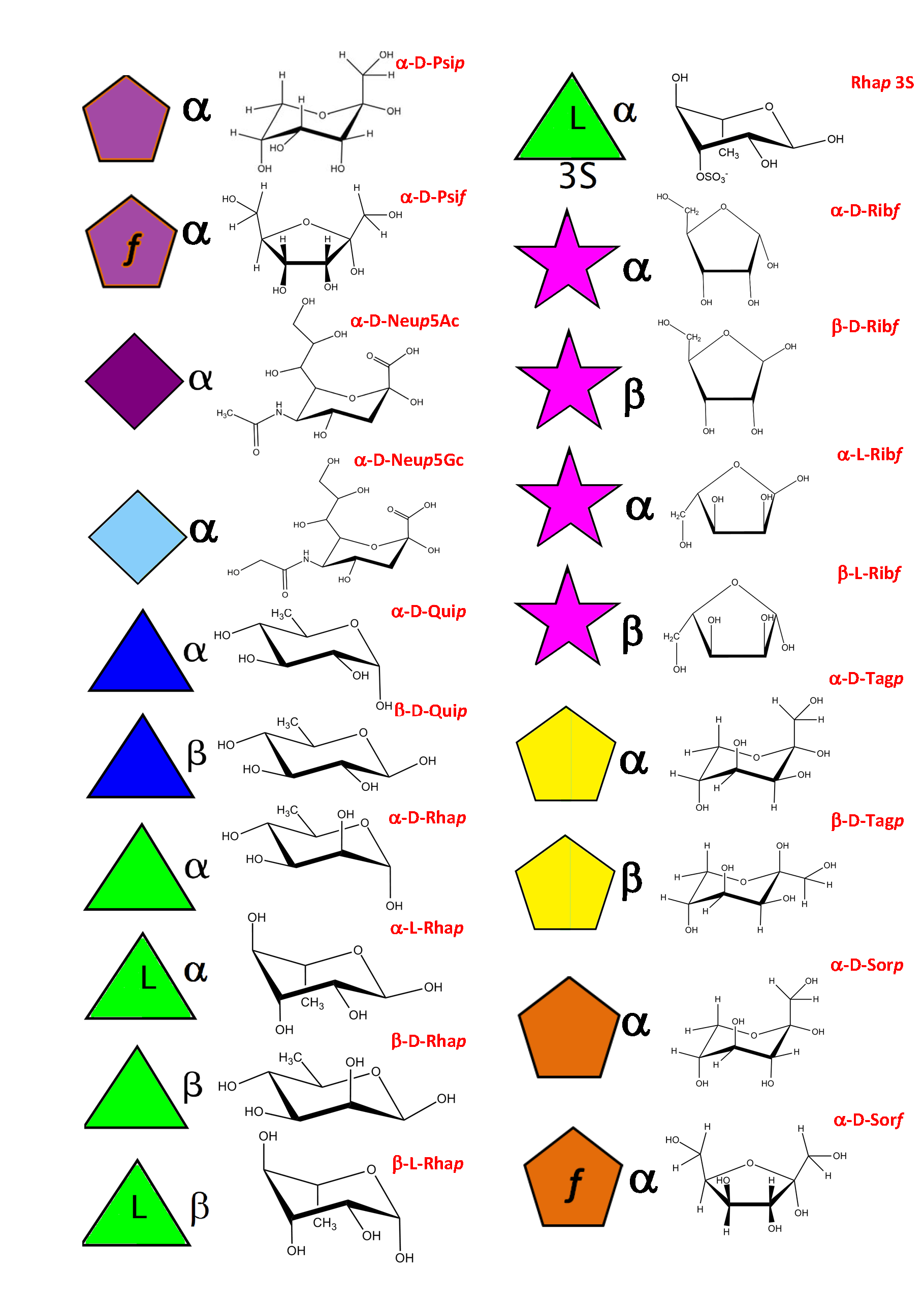
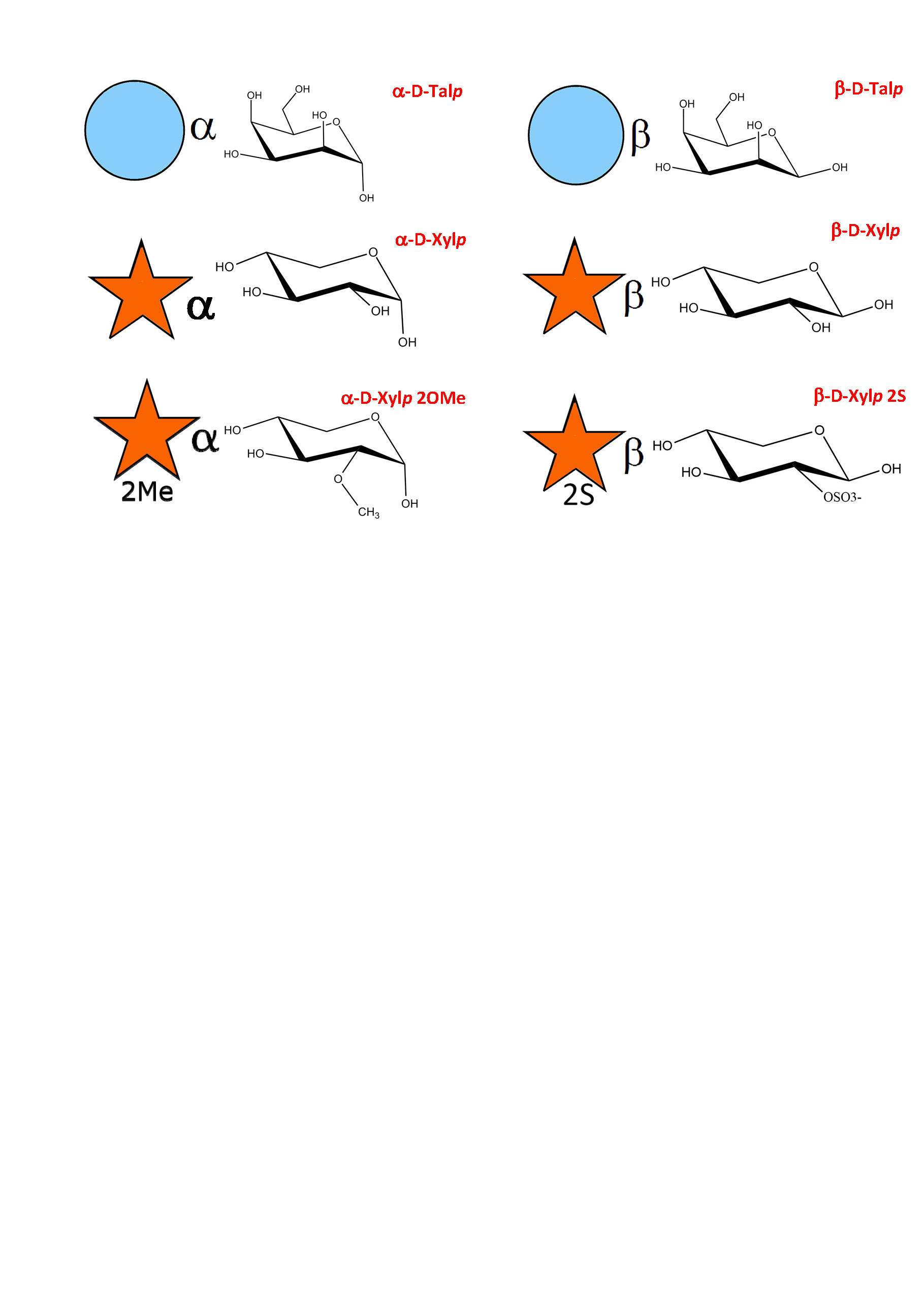
As a result of an extensive search, more than 150 monosaccharides have been identified as the building blocks of the vast majority of oligosaccharides, complex carbohydrates such as “glycan determinants” (blood group antigens, core structures, fucosylated oligosaccharides, sialylated oligosaccharides, Lewis antigens, GPI-anchors, N-linked oligosaccharides, globosides, ….), glycosaminoglycans, plant and algal polysaccharides as well as some bacterial polysaccharides. Each monosaccharide can be assigned a symbolic representation in accordance with the rules set forward, and its cyclic structure represented.
This information is part of the body of an annotated database that contains the three-dimensional structures of the low energy conformations. They have been subjected to systematic conformational sampling to determine their conformational preferences, using molecular mechanics optimization. Whereas most of the monosaccharides exhibit a fairly rigid ring conformation some cases exist where several ring shapes can occur such as in iduronic acid, idose, and all furanosides. In these cases, the low energy conformations are available for each entry. This set of structural information is required to construct three-dimensional structures of complex glycans. www.glyco3d.cermav.cnrs.fr
It is fully compatible with the polysaccharide/glycan three-dimensional builder : Polys 2.0.