Structure and function of C-type lectin receptor DC-SIGN
DC-SIGN is a type II membrane protein with a single CRD, i.e. a type II C-type lectin. It was first cloned and identified as a C-type lectin binding to HIV glycoprotein gp120 (Curtis, B. M, Scharnowske, S, &Watson, A. J. 1992) and later rediscovered and renamed (Geijtenbeek, T. B, Torensma, R, van Vliet, S. et al., 2000) when screening a library of DCspecific monoclonal antibodies that inhibit binding to ICAM-3 (InterCellular Adhesion Molecule-3), the T cells activating adhesion molecule (Geijtenbeek, T. B, Torensma, R, van Vliet, S. J, et al., 2000). DC-SIGN is present at low levels on blood monocytes and as they leave blood to peripheral tissues and differentiate to DCs the expression of this lectin is extremely increased. Finally, following DC maturation and migration to secondary lymphoid tissues, DC-SIGN levels are down-regulated (Geijtenbeek, T. B, Torensma, R, van Vliet, S. J,) . DC-SIGN expression is restricted to certain DC subsets, i.e. dermal and stromal DCs, and it is not expressed on follicular DCs or on skin-resident LCs. Other cell types that express DC-SIGN include macrophages (Granelli-Piperno, A, Pritsker, A, Pack, M, et al., 2005, Lee, B, Leslie, G, Soilleux, E, O’doherty, U, et al., 2001, Soilleux, E. J, Morris, L. S, Leslie, G, et al., 2002, Krutzik, S. R, Tan, B, Li, H, Ochoa, M. T, et al., 2005) and activated B cells (Rappocciolo, G, Piazza, P, Fuller, C. L, et al., 2006). Moreover, there are reports showing the presence of soluble forms of DC-SIGN resulting from alternative splicing (Mummidi, S, Catano, G, Lam, L, et al., 2001, Martinez, O, Brackenridge, S, El-Idrissi, M. E.-A, & Prabhakar, B. S. 2005; Plazolles, N, Humbert, J.-M, Vachot, L, et al., 2011).
The gene encoding DC-SIGN (CD209) is located on human chromosome 19p13.2-3, contains 7 exons and 6 introns, and is closely related to other CLR genes such as DC-SIGNR and CD23. The gene encodes 404 amino acids containing protein (fig. 7) of 44 kDa in molecular weight (Soilleux, E. J, Barten, R, & Trowsdale, J. 2000).
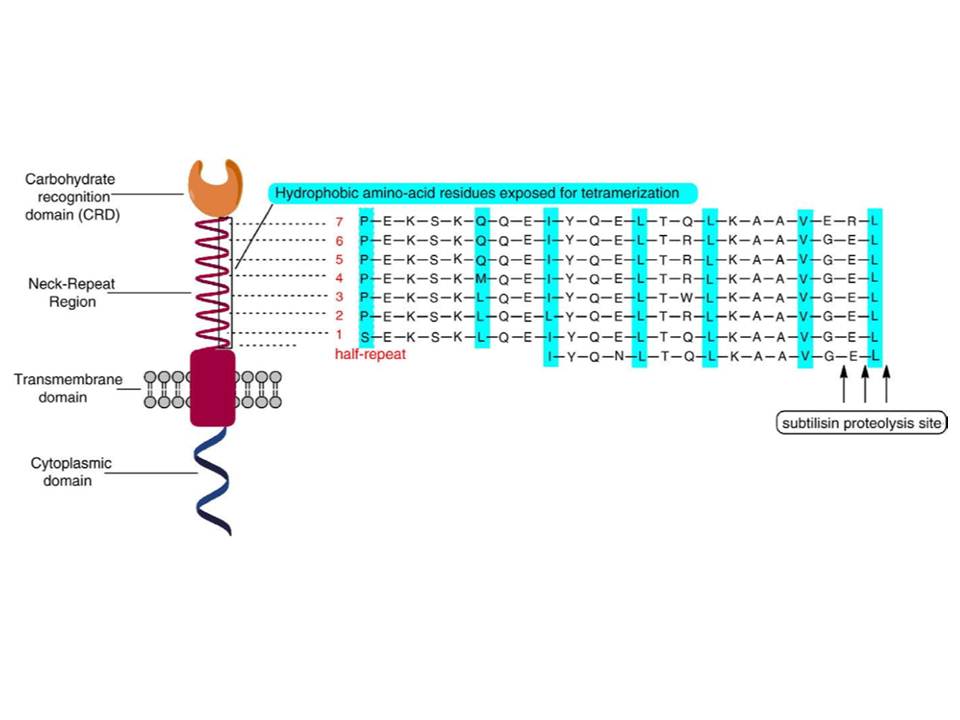
Figure 8.: Domain organization of DC-SIGN molecule.
DC-SIGN CRD: EPN, WND and WMGL motifs are shown in orange, pink and red stick representation; disulphide bridges depicted in yellow sticks and large cyan spheres are the three Ca2+. B, binding of mannose residue in the CRD, the coordination bonds to Ca2+ (cyan sphere) are black solid lines and the Hbonds are represented as black dotted lines. C, Surface representation of DC-SIGN CRD bound to GlcNAc2Man3 (oxygens and nitrogens are in red and blue, carbons of protein and of oligosaccharide are in white and yellow, Ca2+ is green, GlcNAc2Man3 is in stick representation). D, LewisX trisaccharide (upper panel) and a structure of lacto-N-fucopentaose bound to DC-SIGN CRD (lower panel). (A and B are from [73]; C is pdb:1k9i; D is from [Guo, Y, Feinberg, H, Conroy, E, et al., (2004)])
DC-SIGN is composed of four domains : the N-terminal cytoplasmic tail, transmembrane domain followed by the extracellular part composed of a neck region, which contains 7 and a half repeats of 23 amino acid residues, and the C-terminal carbohydrate recognition domain (fig. 7 and 8) (Curtis, B. M, Scharnowske, S, &Watson, A. J. 1992).
DC-SIGN CRD structure and carbohydrate recognition
The Ca2+-dependent CRD of DC-SIGN (fig. 9A) has a typical long-form C-type lectin fold sustained by four disulphide bridges, contains three occupied Ca2+-binding sites (1, 2, and 3) and EPN motif, thus the specificity for mannose group sugars. It binds mannose residues through equatorial 3-OH and 4-OH groups that are coordinated to Ca2+ at site 2, and the overall binding is maintained by the hydrogen bond network formed between distinct sugar groups and amino acid residues in the protein, which can be mediated or not by water molecules (fig. 9B). The interaction with high mannose ligands is not limited to terminal mannose residues and rather involves internal residues that stretch along the extended binding site (fig. 9C) (Feinberg, H, Mitchell, D. A, Drickamer, K, & Weis, W. I. 2001).
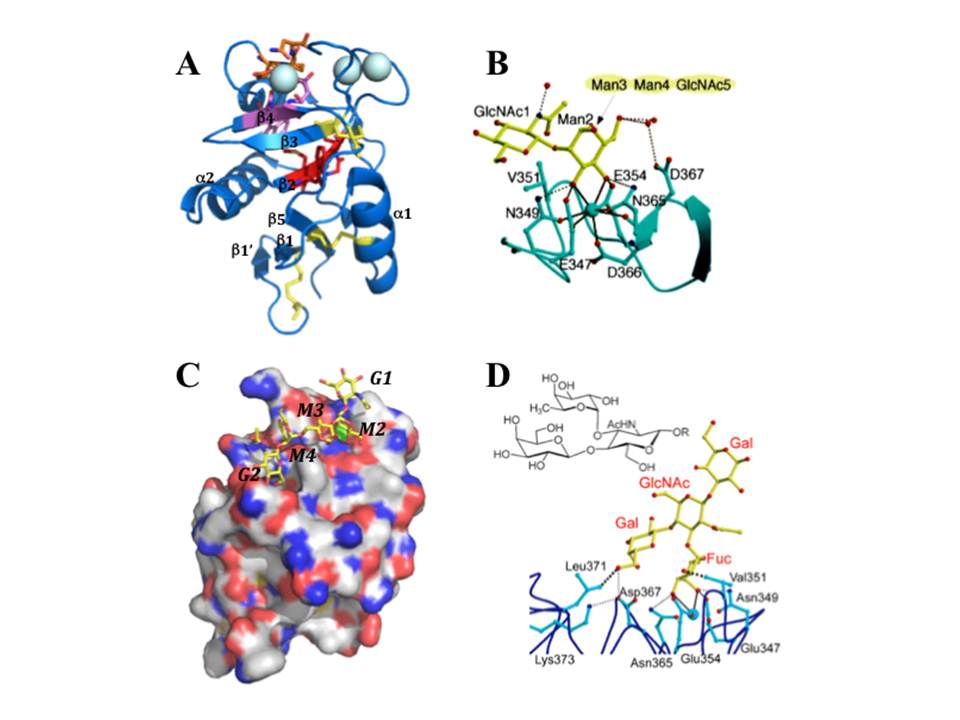
DC-SIGN binds N-acetylglucosamine (GlcNAc) also through equatorial 3-OH and 4-OH groups (Feinberg, H, Mitchell, D. A, Drickamer, K, & Weis, W. I. 2001), and recognizes fucosylated structures, including Lewis-type blood group antigens (Guo, Y, Feinberg, H, Conroy, E, et al., 2004). In this case, fucose residue is bound in Ca-binding site 2 via equatorial 3-OH and axial 4-OH groups, and additional tight van de Waals contacts are formed between 2-OH group of fucose and Val351 residue of the protein (fig. 9D). While GlcNAc residue of LewisX points away from the protein, the terminal galactose residue interacts with the CRD at a secondary binding site through H-bonds and van der Waals contacts with protein side chains. The observed orientation of LewisX bound to DC-SIGN CRD suggests that other fucose-containing oligosaccharides may bind DC-SIGN primarily through fucose in Ca2+-binding site 2 and make additional contacts with the protein at other sites (Guo, Y, Feinberg, H, Conroy, E, et al., 2004).
Oligomeric structure of DC-SIGN
The affinity of the sugars for DC-SIGN CRD is low (KD in mM range). However, the oligomerization of the lectin to a tetrameric form afforded by the neck domain (Mitchell, D. A, Fadden, A. J, & Drickamer, K. 2001) and the heavy glycosylation of its ligands gives a way for high affinity (KD in nM range) interactions due to the avidity phenomenon (fig. 10). The other functions that can be attributed to the neck include the projection of the CRDs further away from the cell membrane for appropriate ligand scanning and binding. The certain positioning of the CRDs due to oligomerization provides additional means of the specificity to the ligands (Frison, N, Taylor, M. E, Soilleux, E, et al., 2003).
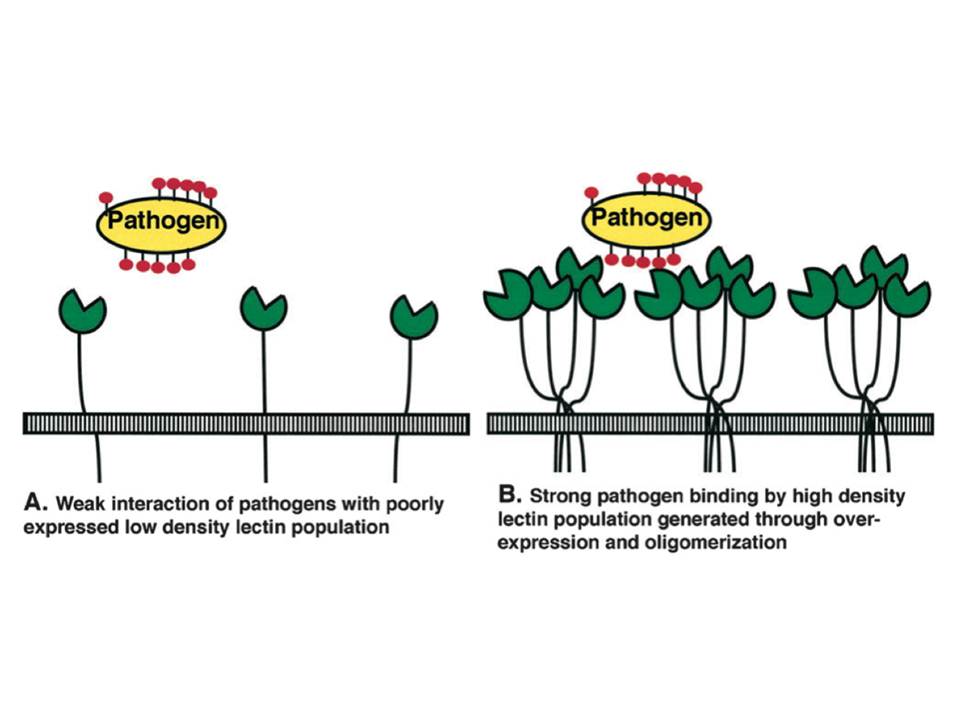
Figure 10 : Schematic representation of lectin/pathogen interaction affinity increase due to the avidity phenomenon
A, Low density of monomeric lectin results in weak interactions.
B, Lectin binding site clustering due to oligomerization and overexpression leads to strong pathogen binding.
Oligomerization of DC-SIGN to tetramers is driven by intermolecular hydrophobic interactions
of amino acid residues of the heptad repeats (fig. 7 and 8) in the neck domains and supported by the lateral salt bridges (fig. 11A) : in the heptads, represented as abcdefg, a and d are hydrophobic, and e and g are charged residues (Lupas, A. 1996).
The studies of truncated DC-SIGN forms with variable length of neck region have shown that at least 6 repeats are needed for the tetramerization, the 5.5 repeats result an equilibrium between tetramers and dimers, and a neck of only 2 repeats leads to equilibrium between dimers and monomers (Feinberg, H, Guo, Y, Mitchell, D. A, et al., 2005). Moreover, complementary studies by the group, have demonstrated that the state and stability of oligomerization is pH-dependent : the decrease of pH induces dissociation of the tetramers into monomers with an approximate mid-point of transition at about pH 5.9. It was also observed that the decrease of pH from mild alkaline (pH 7.9) to slight acidic (pH 5.9) induces the change of the shape of DC-SIGN tetramers from elongated to more compact (Tabarani, G, Thépaut, M, Stroebel, D, et al., 2009).
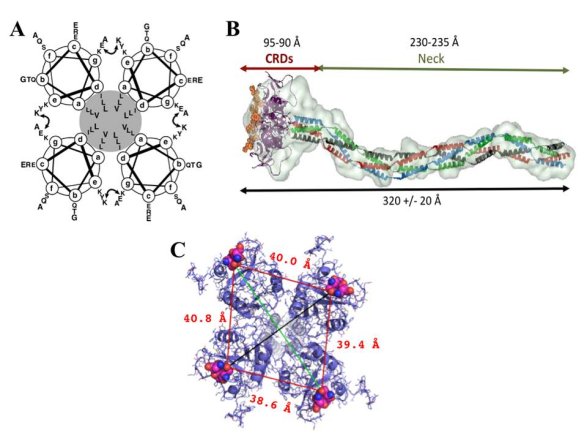
Figure 11. : The structure of the extracellular part of DC-SIGN
A, a schematic representation of the coiled-coil interactions in the neck domain : each of the four circles represents a top-view of an a-helix formed by two 23 amino acid repeats, the hydrophobic core is highlighted in grey, and the curved double arrow represents the putative interchain salt bridge between the glutamic acid (position g) of the first heptad and the lysine residue (position e) of the second one. B, SAXS envelop of DC-SIGN extracellular domain. C, superimposition of DC-SIGN CRDs (pdb:1k9i) into SAXS envelop : the view from the top. The distances between vicinal sugar-binding sites are indicated in red (measurement between the same oxygen atom of Gln147 within EPN motif, depicted as spheres), and diagonal distances in black and green are 51.1 and 61.0 Å, respectively.
The overall length of the extracellular part of the lectin was found to be around 32nm with a certain degree of flexibility observed when binding to the ligand (the length decreases to 28 nm) (Menon, S, Rosenberg, K, Graham, S. A, et al., 2009). This plasticity of the tetramers is likely to be furnished by a flexible CRD-neck junction (Feinberg, H, Guo, Y, Mitchell, D. A, et al., 2005), and allows adapting to the glycan structures on the ligands, thus enabling all CRDs to interact. The global shape of the extracellular part of DC-SIGN revealed by Small Angle X-ray Scattering (SAXS) measurements revealed and confirmed the presumed elongated shape of the lectin with an approximate 40Å spacing between vicinal carbohydrate binding sites (fig. 11B and C) (Tabarani, G, Thépaut, M, Stroebel, D, et al., 2009).
Clustering of DC-SIGN tetramers on the surface of DCs
Regarding the organization of DC-SIGN molecules at the cell membrane level, the lectin is clustered into micro-domains in lipid rafts of the plasma membrane of DCs, and also when ectopically expressed in fibroblasts and other cell types. This clustering occurs even in the absence of the ligands, and the formed micro-domains range from 200nm to 1.5 mm as revealed by different techniques (Cambi, A, de Lange, F, van Maarseveen, N. M, et al., 2004, de Bakker, B. I, de Lange, F, Cambi, A, et al., 2007, Koopman, M, Cambi, A, de Bakker, B. I, et al., 2004).
Moreover, they are remarkably stable in terms of location and diffusion of the receptor within the microdomain, as well as exchange between the micro-domain and its surroundings in the timescale of several minutes (Neumann, A. K, Thompson, N. L, & Jacobson, K. 2008, Itano, M. S, Neumann, A. K, Liu, P, et al., 2011). The recent studies of the nanostructure of DC-SIGN microdomains by super-resolution imaging technique, Blink Microscopy, indicated that DC-SIGN is organized in small 80nm nano-domains (fig. 12), which are randomly distributed on the plasma membrane of DCs and contain as few as 1-3 tetramers as a lower limit, suggesting that other proteins and lipids occupy these nano-domains (Itano, M. S, Steinhauer, C, Schmied, J. J, et al., 2012).
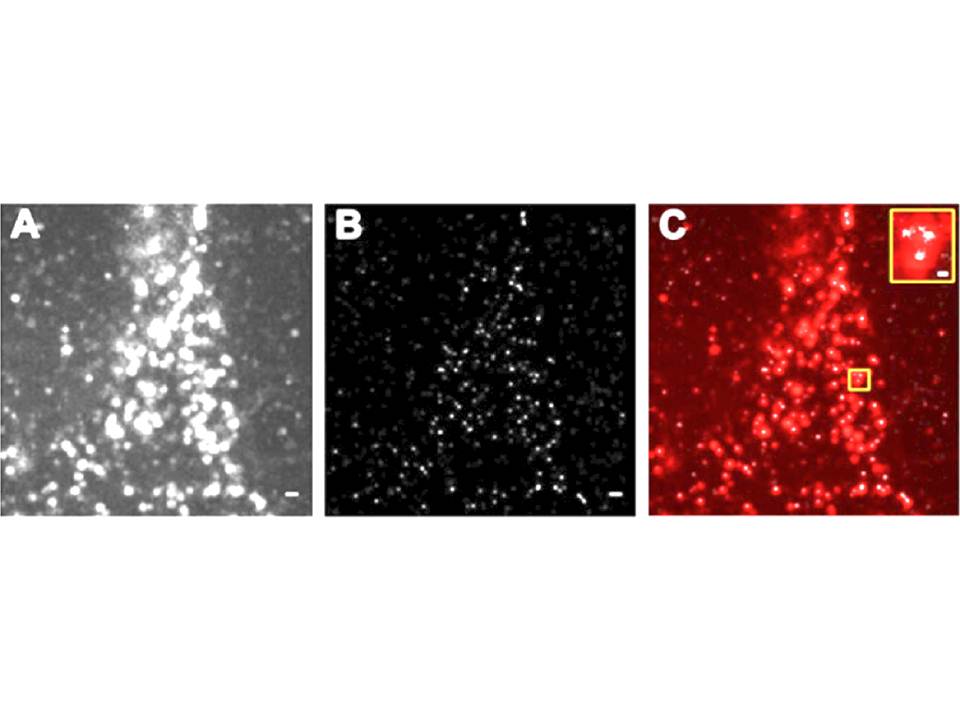
Figure 12 : The nanostructure of DC-SIGN microdomains
A, DC-SIGN expression on fixed DC revealed by diffraction limited total internal reflection fluorescence (TIRF) illumination.
B, Super-resolution Blink image of DC-SIGN expression in the same region as in A. C, superimposition of TIRF image (in red) and Blink image (in white). Scale bars, 500 nm ; scale bar inset, 100 nm.
The mechanism of DC-SIGN clustering on the membrane was investigated using truncated forms of DC-SIGN and the influence of distinct DC-SIGN parts for the formation and stability of micro-domains was examined (Liu, P, Wang, X, Itano, M. S, Neumann, A. K, et al., 2009). These studies indicated that the driving force of micro-domain formation is the saccharide binding to CRD, since CRD-depleted DC-SIGN or mutants of a single amino acid required for Ca2+-coordination at sugar binding site showed a diffused expression over all the membrane with no visible micro-domains and high mobility. The neck domain appeared to be responsible for the stability of the micro-domains, as in case of DC-SIGN depleted of tandem repeats the micro-domains were dynamic, often moving on membrane from one position to another, and DC-SIGN within these domains was able to exchange with other DC-SIGN molecules from the surrounding membrane. By contrast, the removal of whole cytoplasmic domain and N-glycosylation site at Asn80 had no significant effect on micro-domain formation and stability suggesting that the clustering mechanism is not dependent on cytoskeletal structures, despite the observation that DC-SIGN constitutively binds to scaffold proteins LSP1, KSR1 and CNK (Gringhuis, S. I, den Dunnen, J, Litjens, M, et al., 2009).
The finding that CRD is essential for the clustering of DC-SIGN on the membrane suggests several mechanisms : (1) CRD directly binds to polysaccharides of the extracellular matrix ECM), e.g. glycosaminoglycans or glycosyl moieties of ECM proteins (fig 13A) ; (2) CRD directly binds to trans-membrane proteoglycans that link to the ECM (fig 13B) ; (3) glycosylated transmembrane adaptor proteins (TRAPs), which are directly or indirectly linked to the membrane-apposed cytoskeleton and have cis interactions with DC-SIGN, serve to stabilize the micro-domains (fig 13C).
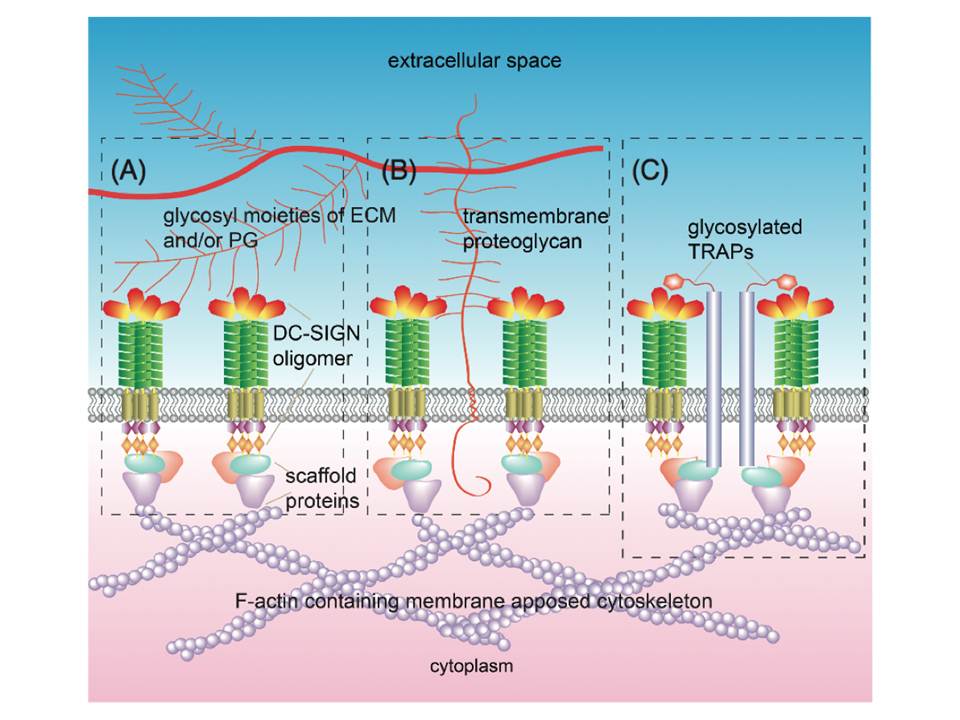
Figure 13 : Possible mechanisms of DC-SIGN microdomain formation and stability
Microdomain formation facilitated and stabilized through DC-SIGN CRDs interactions with ECM components (A),transmembrane proteoglycans that are linked to ECM components (B), and/or glycosylated TRAPs, having cis interactions with DC-SIGN, link the receptor to the cytoskeleton (C).
Such clustering of DC-SIGN into micro-domains further contributes to the avidity of DC-SIGN interactions with its ligands. Furthermore, it has been proposed that such clustering may modulate DC-SIGN specificity by favoring carbohydrates with a certain density and spacing (Cambi, A, Koopman, M, & Figdor, C. G. 2005).
Ligand recognition by DC-SIGN
Endogeneous and exogeneous ligands of DC-SIGN
DC-SIGN has been discovered as a C-type lectin of DCs that binds to the T cells adhesion molecules ICAM-3, and this interaction is believed to mediate transient, antigen nonspecific adhesion of DCs with T cells (fig. 14A), leading to effective T cell receptor engagement and screening of the MHC-peptide complexes (Geijtenbeek, T. B, Torensma, R, van Vliet, S. J, et al., 2000). DC-SIGN binds ICAM-3 through the recognition of the N-glycans of high mannose-type oligosaccharides and, depending on the cell population, Lewis X residues (Bogoevska, V, Nollau, P, Lucka, L, et al., 2007). ICAM-2 that is expressed on endothelial cells was identified as another endogenous counterstructure of DC-SIGN (Geijtenbeek, T. B, Krooshoop, D. J, Bleijs, D. A, et al., 2000). The interaction of these two molecules resists shear stresses and is involved in DC adhesion, tethering, and rolling along the ICAM-2 expressing surfaces (fig. 14A). DC-SIGN binding to ICAM-2 was shown to be strictly glycan-specific and LewisY structures on ICAM-2 were identified as the glycans possibly mediating this interaction (García-Vallejo, J. J, van Liempt, E, da Costa Martins, P, et al., 2008). Additionally to the adhesion molecules ICAM-2 and ICAM-3, DC-SIGN binds to Lewis X expressed on b2-integrin Mac-1 and CEACAM1 on neutrophils (PMNs), and thus drives the interaction between DCs and neutrophils (fig. 14B) (van Gisbergen, K. P. J. M, Sanchez-Hernandez, M, Geijtenbeek, T. B. H, & van Kooyk, Y. 2005, van Gisbergen, K. P. J. M, Ludwig, I. S, Geijtenbeek, T. B. H, & van Kooyk, Y. 2005), and by binding to LewisX and LewisY sugars recognizes carcinoembryonic antigen (CEA) overexpressed on majority of colorectal cancer cells (van Gisbergen, K. P. J. M, Aarnoudse, C. A, Meijer, G. A, et al., 2005). DC-SIGN also recognizes human semen clusterin and this binding interfers HIV capture by DCs (Sabatte, J, Faigle, W, Ceballos, A, et al., 2011).
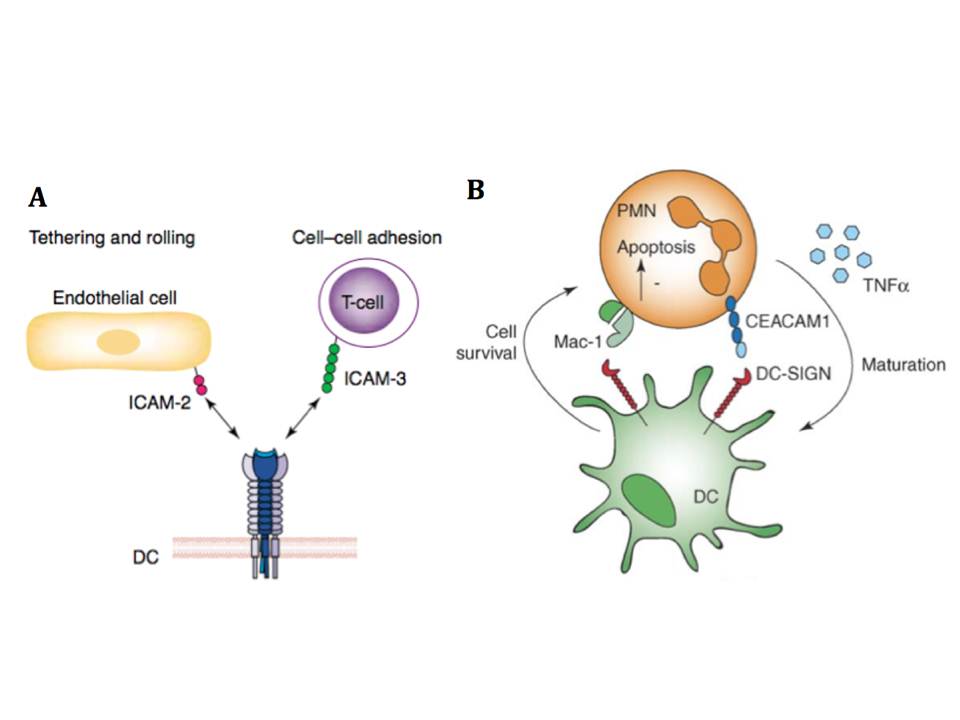
Figure 14 : The physiological role of DC-SIGN and endogeneous ligands
A, DC-SIGN promotes transmigration of DCs from the blood vessels into lymphoid tissues by interacting with ICAM-2 expressed on endothelial, and mediates immunological synapse initiation by binding to ICAM-3 on T cells (from [98]). B, DC-SIGN binds Mac-1 and CEACAM on PMNs and induces PMN survival, while PMNs can induce DC maturation
DC-SIGN also functions as a PRR since it is capable to recognize the glycans expressed on various pathogens. It has been shown that DC-SIGN is implicated in antigen processing and presentation to T cells (Engering, A, Geijtenbeek, T. B. H, van Vliet, S. J, et al., 2002). Initially, DC-SIGN was found to bind HIV-1 through interaction with envelope glycoprotein gp120, but soon it appeared that this lectin also captures many other dangerous pathogens (table 1.2). Unfortunately, this recognition does not lead to infection clearance, but rather the pathogens exploit DC-SIGN to escape immunity and facilitate their invasion and infective processes.
Ligand-induced DC-SIGN signaling
The ligand binding to DC-SIGN triggers endocytosis and cellular signaling, which requires simultaneous TLR signaling and is dependent on the nature of the ligand. The cytoplasmic domain of DC-SIGN contains di-leucine (LL) and triacidic cluster (EEE) and other internalization motifs, which direct the DC-SIGN-bound ligands into late lysosomes or MHC class II positive endosomes (Engering, A, Geijtenbeek, T. B. H, van Vliet, S. J, et al., 2002). However, this process is ligand-dependent since HIV-1 virions bound to DC-SIGN are directed to early endosomes (Kwon, D. S, Gregorio, G, Bitton, N, et al., 2002). DC-SIGN-mediated internalization has been shown to occur via clathrin-coated pits and requires membrane cholesterol and dynamin Cambi, A, Beeren, I, Joosten, B, et al., (2009), but it is worth to note that ligands, as antibodies, binding to DC-SIGN neck region induce clathrin-independent internalization that directs ligands to early endosomes (Tacken, P. J, Ginter, W, Berod, L, et al., 2011).
The cytoplasmic tail of DC-SIGN also contains an incomplete ITAM indicating the lectin’s signaling capability (104). Indeed, the final outcome of DC-SIGN triggering is the gene transcription by NF-kB (reviewed in Geijtenbeek, T. B. H & Gringhuis, S. I. (2009), Geijtenbeek, T. B. H, den Dunnen, J, & Gringhuis, S. I. 2009), despite that DC-SIGN is considered ITAM/ITIM independent CLR. NF-kB is an important transcription factor composed of several subunits and activated by several classes of PRRs following pathogen recognition. It regulates the expression of genes encoding costimulatory molecules, cytokines and chemokines. However, the proper induction of gene transcription requires NF-kB translocation to nucleus, and this cannot be accomplished by triggering DC-SIGN alone – the prior activation of NF-kB by TLR signaling is necessary. Thus, DC-SIGN modulates the NF-kB activity and the cross-talk between DC-SIGN and TLR signaling gives the final immune response.
Mannose-containing ligand binding to DC-SIGN induces the downstream effector kinases, which further leads to Ser/Thr protein kinase RAF1 activation (fig 15). The activated RAF1 then phosphorylates p65 subunit of NF-kB, which makes it a target for subsequent acetylation and finally increases its activity (prolongs its nuclear activity, enhances transcriptional rate and increases binding affinity to DNA) on Il6, Il8, Il12a and Il12b promotors, and particularly on Il10 promotor, which strongly increases anti-inflammatory IL-10 production. Although RAF1 activation is followed by DC-SIGN triggering and does not require TLR signaling, p65 cannot be activated by DC-SIGN alone – DC-SIGN signaling only modulates p65 activation when it has been already translocated to nucleus (reviewed in Geijtenbeek, T. B. H, den Dunnen, J, & Gringhuis, S. I. 2009, van den Berg, L. M, Gringhuis, S. I, & Geijtenbeek, T. B. H. 2012).
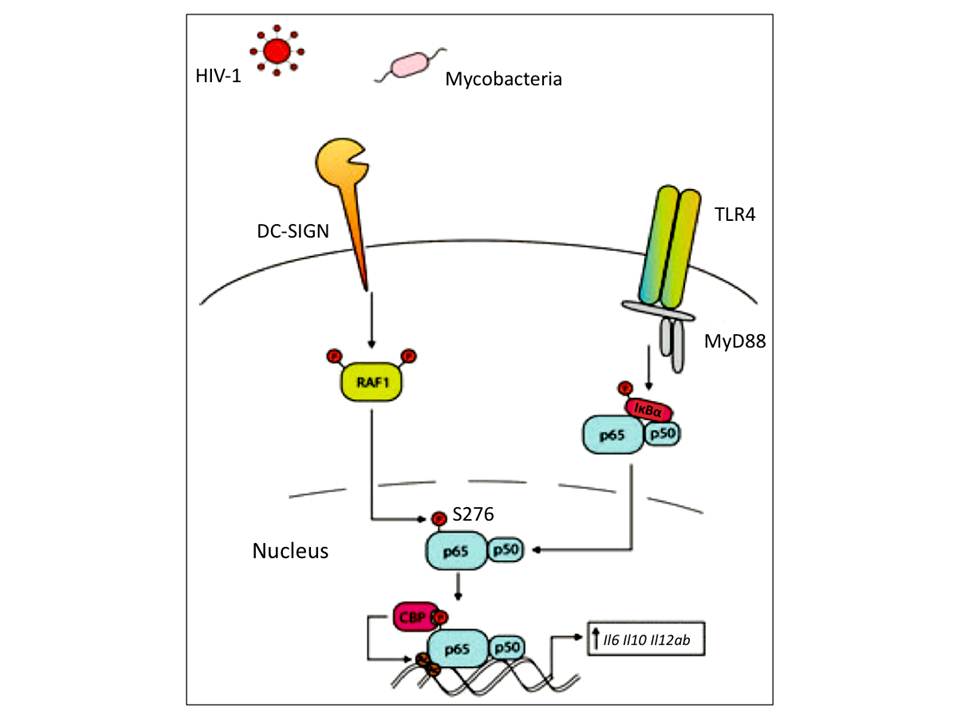
There is very limited knowledge about DC-SIGN signaling in response to fucose-containing pathogen binding, and it comes only from LewisY positive and negative H. pylori infection studies and examinations of immune responses to soluble egg antigens of the parasite Schistosoma mansoni (reviewed in Geijtenbeek, T. B. H, den Dunnen, J, & Gringhuis, S. I. 2009). Only LewisY+ H. pylori interacts with DC-SIGN, which induces high up-regulation of only anti-inflammatory IL-10 production. Such a response is significantly different from responses to LewisY- H. pylori strain as well as to mannose-containing pathogens that induce up-regulation of both pro-inflammatory cytokines such as IL-6 and IL-12, and anti-inflammatory IL-10.
Thus the pathogens evolved to exploit DC-SIGN signaling for their infection promotion. The cross-talk between DC-SIGN and TLR signaling also may explain why DC-SIGN recognition of endogenous ligands does not lead to DC maturation and cytokine production : since there is no simultaneous activation of PRRs by pathogens to induce NF-kB activation, DC-SIGN triggering alone does not induce immune responses (van den Berg, L. M, Gringhuis, S. I, & Geijtenbeek, T. B. H. 2012).
