In 1884, the Danish bacteriologist, Hans Christian Gram, while trying to set up a protocol to stain bacteria for observation under the microscope, developed a technique, which became fundamental to discriminate bacteria according to the composition of their cell wall. Heat-fixed bacterial cells are first treated with a purple dye, gentian violet, which penetrates through the cell wall and plasma membrane, thus staining the cytoplasmic compartment. The addition of iodine, which binds to the violet dye through an ion pair, traps it into the cell. When a decolorizer such as ethanol is added to the fixed cells, two behaviors can be observed, either (i) the purple color is retained, or (ii) the purple color is washed out and a secondary staining with safranin or fuchsin is necessary to give decolorized bacteria a pink or red color for visualization. The permeability of the cells to the decolorizer and the washing-out of the primary dye-iodine complex depend on the architecture of the cell wall. The latter contains an essential and ubiquitous peptidoglycan -or murein- layer, which results from the polymerization of β-1,4-linked N-acetyl-glucosamine (GlcNAc) and N-acetyl-muramic acid (MurNAc) disaccharide units, cross-linked by short peptide stems. If differences in the chemical structure of the individual motifs are rather small, the thickness of the peptidoglycan layer nevertheless drastically differs within bacteria. Bacteria with a thick peptidoglycan layer tend to retain the primary dye of the Gram protocol, while a thin peptidoglycan layer favors the washing-out of the dye-iodine complex. Nevertheless, peptidoglycan is not the exclusive component of the cell wall. Bacteria that give a negative staining in the Gram protocol tend to surrender their thin peptidoglycan layer with a lipid-containing outer-membrane, which is destabilized and washed-out by the addition of the ethanol decolorizer. In contrast, bacteria that give a positive staining in the Gram protocol present a surface layer that tends to be dehydrated upon the ethanol treatment. Some bacteria, such as the genera Actinomyces, Corynebacterium or Mycobacterium, yield a Gram-variable pattern with this protocol. A detailed presentation of the cell wall of typical Gram-positive and Gram-negative bacteria follows, completed by a description of some of these specific Gram-indeterminate bacteria.
Gram-positive bacteria
Gram-positive bacteria encompass organisms such as the rod-shaped model Bacillus subtilis, the spheroid Staphylococcus aureus, or the ovococcus Streptococcus pneumoniae. Because of their thick peptidoglycan layer (Figure 2), which prevents washing of the gentian violet dye, Gram-positive bacteria appear in purple with Gram-staining. Their wall is further characterized by a single membrane (plasma or inner-membrane) and the presence of different glycopolymers, that are connected either to the peptidoglycan or to lipids of the plasma membrane (for a review, see Weidenmaier & Peschel, 2008). These cell wall glycopolymers thread through the peptidoglycan layers towards the bacterial cell surface, where they can shape physicochemical surface properties and biofilm formation, mediate interaction with host receptors or binding to phages, and initiate innate host defenses and inflammation, T-cell or complement activation, or opsonization. In addition, the surface of Gram-positive bacteria can be covered with protective surface structures, such as capsular polysaccharides or surface layer (S-layer) proteins, which are highly variable among bacterial species and modulate all the previously described activities (see for example Eberhardt et al., 2012 for the attachment of capsular polysaccharides in S. pneumoniae).
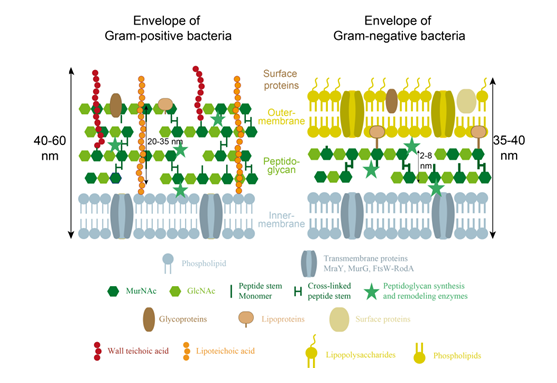
The peptidoglycan-anchored cell wall glycopolymers are usually covalently linked to the peptidoglycan N-acetylmuramic acid through a phosphorylated disaccharide composed of N-acetylglucosamine and another sugar. The glycopolymer itself can be zwitterionic, as in most teichoic acids, anionic as in most teichuronic acids, or neutral when other sugars such as mannose or galactose are involved. Teichoic acids, as in S. aureus Xia et al., 2010, are generally formed by repeats of polyglycerol and/or polyribitol phosphate residues bound by phosphodiesters. Their zwitterionic properties come from the negative charge of their phosphate groups in physiological conditions, balanced with the amino extremities of the D-alanine polyol elements. When D-alanylation is replaced by glycosylation as in B. subtilis strain 168 or VKM 501 Bougault et al., 2012, the teichoic acid becomes anionic. In cases of phosphate starvation, teichuronic acids are produced instead of wall teichoic acids, as in the case of the B. subtilis strain 168 Fritz & Mascher, 2014. The anionic properties of this cell wall glycopolymer then result from the negative charge on the carboxylate group of the glucuronic acid. Alternatively, the secondary cell wall polysaccharides of the Bacillus cereus group of Gram-positive bacteria, that includes Bacillus anthracis, B. cereus and Bacillus thuringiensis strains, usually provide a neutral cell wall glycopolymer Choudhury et al., 2006.
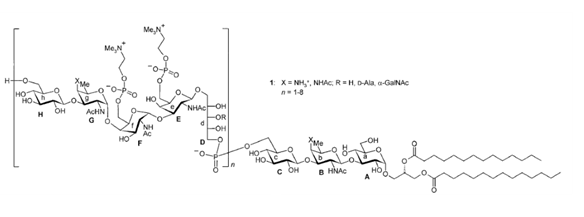
The structure of the membrane-anchored glycopolymers is usually less diverse than their peptidoglycan-linked analogs. They usually consist of lipoteichoic acids containing glycerol-phosphate repeating units that are connected to lipids through a glycerol-disaccharide anchor. However, more complicated lipoteichoic acid structures have also been described, such as the ribitol tetrasaccharide motif of Streptococcus pneumoniae Klein et al., 1996, shown in Figure 3.
Gram-negative bacteria
Gram-negative bacteria include the well-known Escherichia coli or the crescent-shaped model Caulobacter crescentus. The lighter color of these bacteria by Gram coloration is due to the inability of these organisms to retain the purple gentian violet dye during the washing step. Indeed, they possess a much thinner peptidoglycan layer than Gram-positive bacteria, but this is enclosed in the periplasmic space between an inner (cytoplasmic) and an outer membrane (Figure 2, Silhavy et al., 2010). The outer-membrane is an asymmetrical membrane composed of phospholipids and glycolipids (or LipoPolySaccharides ; LPS), at the inner and outer leaflets, respectively. The latter molecules consist of a lipid A anchor linked to a core oligosaccharide, that can be extended with an O-antigen polysaccharide of variable length which protects the cell against macrophages and complement system from the innate immune response Putker et al., 2015. Lipopolysaccharides form an almost impermeable layer at the surface of Gram-negative bacteria that acts as a protective barrier against antibiotics and other antimicrobial molecules, and more generally hydrophobic molecules.
Embedded in the outer-membrane, proteins can also be found. With a few exceptions, they can be divided in two classes, lipoproteins and β-barrel proteins Silhavy et al., 2010. The former contain a lipid moiety generally covalently linked to a cysteine residue and are mainly localized in the inner leaflet of the membrane. One of them, the highly abundant Braun’s lipoprotein (also known as Lpp), is actually essential in E. coli for cell wall integrity as it is covalently linked to peptidoglycan to tether the outer membrane to the murein layer. Alternatively, β-barrel proteins are transmembrane proteins that allow the passive diffusion of small molecules, such as sugars and amino acids, through the outer membrane (porins), or that function as gated channels for the transport of high affinity ligands such as Fe-chelates or vitamins. Some additional proteins, usually specific to the bacterial species can also be found in the outer membrane.
The periplasmic space delimited by the inner- and outer-membranes not only contains the peptidoglycan polymer, but also a high density of various proteins and chaperones, which are involved in the biosynthesis or maturation of the peptidoglycan and cell envelope polymers, but also in sugar and amino acid transport and chemotaxis (for a review see Silhavy et al., 2010. Secretion systems for example recruit a number of proteins from the cytoplasm to the outer-membrane, that associate to form a dynamical trans-envelope assembly which can release proteins, DNA or toxins in the medium or to prokaryotic or eukaryotic cell (for a review, see for example Costa et al., 2015).
The cell wall of bacteria with intermediate Gram-staining
Although most of bacteria can be classified either as Gram-positive or as Gram-negative, some cases are more complex. For instance, the notorious bacillus Mycobacterium tuberculosis is hardly stained by Gram coloration. The cell wall of mycobacteria and other corynebacteria is composed of five layers (Figure 4), which are the plasma membrane, the granular layer, the inner wall zone, the medial wall zone and the mycomembrane, when starting the description from the inside to the surface of the cell Zuber et al., 2008.
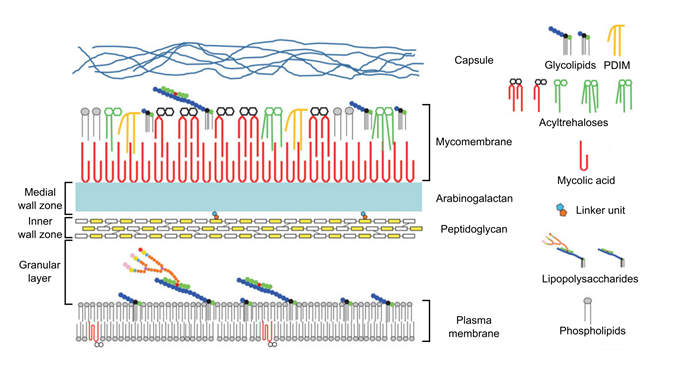
The plasma membrane is very similar to that of Gram-positive and Gram-negative bacteria. The approximately 8.5-nm thick granular layer is similar to that detected in the Gram-positive Streptococci and is apparently bound to the plasma membrane, while it is absent in Gram-negative bacteria. This is one of the reasons why, despite its response to the Gram-staining, mycobacteria and corynebacteria are often classified within the Gram-positive family. The 15-nm thick inner wall zone is of low density and mainly contains peptidoglycan, while the 7-nm thick medial wall zone is of intermediate density and contains peptidoglycan covalently bound to arabinogalactan, a glycopolymer composed of 1,4-linked galactose and arabinose. The arabinogalactan is in turn covalently bound to mycolic acids, which are very-long chain of α-branched and β-hydroxylated fatty acids that are specific to the Corynebacterineae. The mycolic acids interact with various lipids and glycolipids to form the mycomembrane, which is highly impermeable, similar to the outer-membrane of Gram-negative bacteria Daffé, 2015. Embedded in this membrane, porins can also be found, which form specific hydrophobic pores in this structure. The outermost layer of the cell envelope represents a capsule-like material mainly composed of polysaccharides and proteins, and eventually in some specific cases specialized glycolipids.
Corynebacterieae are not the only example of bacteria with a non conventional cell wall. Until 2014, it was supposed that intracellular pathogens Chlamydiae and aquatic Planctomycetes phyla did not possess any peptidoglycan layer, although the former were known to be sensitive to antibiotics targeting its synthesis. Using peptidoglycan probes and cryo electron-microscopy, new studies however revealed the presence of a murein layer enclosed between two membranes in some of these organisms (see Pilhofer et al., 2013;Liechti et al., 2014 for Chlamydiae and Jeske et al., 2015van Teeseling et al., 2015 for Planctomycetes). These results should trigger a renewal of interest for species from these phyla, as they are currently the only known bacteria with peptidoglycan synthesis machineries totally independent of the usually essential FtsZ division protein.
Peptidoglycan-less bacteria
Interestingly, some bacterial organisms are totally devoid of any bacterial cell wall, therefore lacking peptidoglycan. The most common ones known to date are from the Mollicutes phylum. Indeed, bacteria from their most studied genus, Mycoplasma, are among the tiniest organisms (0.2 to 0.3 µm) and have, excluding viruses, the smallest genome (between 0.6 and 1.35 Mbp), presumably due to a loss of their genes throughout evolution. As a consequence, they are lacking almost all – if not all – the required genes for peptidoglycan synthesis enzymes. Mycoplasma are bound by a single plasma membrane that is made of choline-containing phospholipids, among which phosphatidylcholine and sphingomyelin, cardiolipin and phosphatidylglycerol Park et al., 2013. The absence of rigid cell wall induces an extreme sensitivity to osmotic pressure, which is the reason why these organisms are depending on the adhesion to the surface of a eukaryotic host cell. More recently, three spherical and chemoheterotrophic Gram-negative bacteria were isolated, affiliated to the genus Cerasicoccus within the phylum ’Verrucomicrobia’, and were reported to lack peptidoglycan Yoon et al., 2010. This suggests that more peptidoglycan-less bacteria may be discovered in the future.
Despite the primary importance of peptidoglycan in bacteria with a cell wall, it has been shown that some Gram-positive and Gram-negative cells can also survive without this structure in a state called L-form. Indeed, such wall-free bacteria can be obtained from the inhibition of cell wall synthesis in osmoprotective conditions Allan, 1991 Leaver et al., 2009. The generated organisms lose their initial shape, become round, and lose their rigidity. Although still little information is known about bacterial L-forms, it is suggested that they might be implied in diseases Allan et al., 2009 and it has been hypothesized that they could be a relic of an antique shape existing before the apparition of sacculi Errington, 2013.
