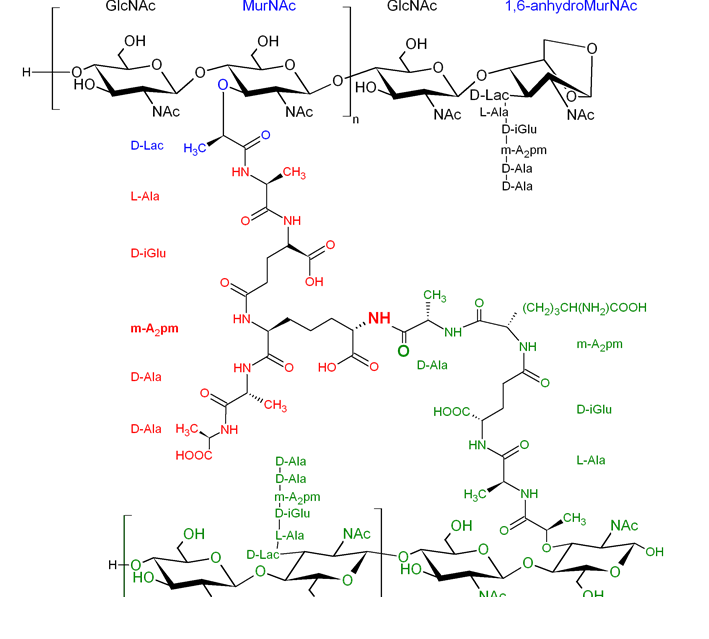
Chemical composition
In all bacteria, the glycan strands of peptidoglycan are composed of alternating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc, a variant of GlcNAc with a D-lactate attached to the C-3 by an ether bond) as shown in Figure 8. Generally, the obtained strands are terminated at the reducing end by a 1,6-anhydroMurNAc residue, in which the C-1 and C-6 of the sugar backbone are bound through an ether linkage. The unique features of this residue are used to quantify the glycan chain average length. The peptide stems are covalently linked to the glycan strands with an amide bond to the carboxyl carbon of the D-lactyl group of the MurNAc. While the chemical composition of the glycans is highly conserved across species, the peptide sequence is more diverse and has the particularity to involve D-amino-acids. In the first position from the lactyl group, an L-alanine (L-Ala) is usually found, and in rare exceptions a Gly or an L-Ser residue. A D-isoglutamic acid (D-iGlu) follows, which is sometimes amidated in Gram-positive bacteria to yield a D-isoglutamine (D-iGln), as in S. pneumoniae Zapun et al., 2013. The γ-carbon of this residue is connected to a third amino acid, which is most probably the most variable within the stem peptide Vollmer, Blanot et al., 2008. In most Gram-negative species, Gram-positive bacteria of the genus Bacillus and mycobacteria, the third amino acid is usually the meso-diaminopimelic acid (m-A2pm), while in most-Gram positive species it is usually an L-lysine (L-Lys). The peptide stem is finally terminated by two D-alanines (D-Ala), although different D-amino acids can be found either naturally or by addition of specific amino acids in the growth medium.
Cross-links at the peptide stems
The percentage of cross-linking in peptidoglycan can be determined by a combination of HPLC and MALDI-TOF MS. Analyzes can be complemented by MS/MS experiments on the individual muropeptides to validate the structure of each individual species by fragmentation. These analyzes can also be complemented by Nuclear Magnetic Resonance Spectroscopy.
Two main cross-linking strategies have been identified in bacteria up-to-date. The most widespread strategy yields to a 4→3 cross-link, by connecting the amino group of the 3rd residue (m-A2pm or L-Lys) of the acyl-acceptor stem peptide to the carbonyl moiety of the 4th amino acid (D-Ala) of the acyl-donor stem peptide (Figures 8 and 9). This reaction is catalyzed by a D,D-transpeptidase enzyme (among which the Penicillin-Binding Protein (PBP) enzymes), which utilizes disaccharide-pentapeptides as substrate. In this reaction, the D-Ala in position 5 of the acyl-donor peptide stem is thereafter eliminated. An alternative strategy was evidenced in an in vitro selected ampicillin-resistant Enterococcus faecium strain Mainardi et al., 2000 and involves a 3→3 cross-link. In this case, transpeptidation occurs between the amino group of the 3rd residue (m-A2pm or L-Lys) of the acyl-acceptor stem peptide and the carbonyl moiety of the 3rd amino acid (m-A2pm or L-Lys) of the acyl-donor stem peptide. This reaction is catalyzed by L,D-transpeptidases Mainardi et al., 2005 and specifically requires disaccharide-tetrapeptide substrates (Figure 9). Although this cross-linking mode is usually in minority, it can become a major strategy in some species, as reported for M. tuberculosis (80% ; Lavollay et al., 2008). From computer modeling, it was postulated that 3→3 cross-links might induce stiffness in the peptidoglycan, due to the replacement of the D-Ala in position 4 by the m-A2pm donor in position 3 de Pedro & Cava, 2015. Nevertheless measurements by solid-state NMR on 3→3 and 4→3 cross-linked peptidoglycan seem to indicate that the polymer flexibility remains mainly unaffected in the μs-to-ns time-scale dynamical regime Gansmüller et al., unpublished results. The D,D- and L,D-transpeptidase enzymes are responsible for these 4→3 and 3→3 cross-links, respectively.
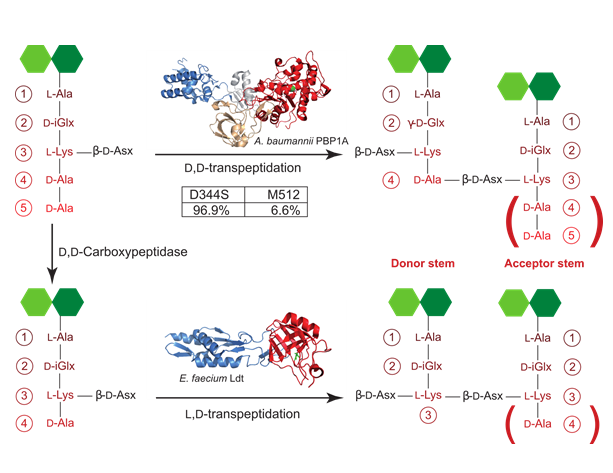
Additionally to dimeric species, trimers and tetramers can be detected in the analysis of muropeptides. This is the case if the cross-linking involves one or two more glycan chains. In the case of a trimer, the 4th residue of the acceptor stem in the constitutive dimer can play the role of donor in a new transpeptidation reaction and make a cross-link with the third residue of another acceptor peptide stem. Similarly for a tetramer, the D-Ala of the trimer acceptor peptide stem is connected to a neighboring peptide from another glycan strand. In both cases, these multimerizations produce structural constraints forcing the third and fourth strands to be on distinct plans from the initial peptidoglycan surface de Pedro & Cava, 2015. One could suggest that such knots, if accumulated locally, are likely to modify peptidoglycan physical properties and would favor a rather disordered net. However, in most species studied until now, trimers are present in limited quantities and tetramers are even much rarer. As an example, trimers and tetramers were only found in less than 10% and 0.25% of the cross-links in E. coli, respectively Glauner et al., 1988. It is hard to determine whether these amounts are sufficient to impact the sacculus physical properties, especially for the tetramers.
On the contrary to Gram-negative bacteria, it is not rare in Gram-positive organisms to find cross-linking through a bridge. This peptide bridge can be composed of one to seven amino acids and various amino acids can be encountered, which can be connected through their side chains instead of their backbones as the β-D-Asx in E. faecium (Figure 9). For example, in S. aureus, the peptide bridge in the cross-link is made of a succession of five glycines. Such a bridge is also required in the peptidoglycan of corynebacteria, which is specific in cross-linking the α-carboxyl carbon of the D-iGlu in position 2 of one peptide stem (acyl acceptor) to the carbonyl carbon of the D-Ala at position 4 of another peptide stem (acyl donor). Of note, the cross-link in these species is a very unusual 2→4 cross-link, whose formation is nevertheless catalyzed by PBPs Vollmer, Blanot et al., 2008for a review.
Cross-linking may also not be uniform across the peptidoglycan as in S. pneumoniae Bui et al., 2012, which contains direct cross-links and cross-linking through L-Ser-L-Ala or L-Ala-L-Ala bridges. Furthermore, peptide cross-linking of the glycan strands is not a process, which necessarily happens as soon as the murein is synthesized. Actually, new peptidoglycan has been shown to be less cross-linked than “old” peptidoglycan Burmant & Park, 1983de Pedro & Schwarz, 1981. Similarly, the degree of cross-linking can also be impacted by the bacterial stage of growth Pisabarro et al., 1985 and environmental conditions.
Modification of the peptidoglycan
The peptidoglycan net obtained after polymerization of the glycan strands and cross-linking of the peptide stems rarely remains unaltered. Several chemical modifications have been reported in the glycan strands. These are usually situated on the C-2 and C-6 of MurNAc and/or GlcNAc (Figure 10 Vollmer, 2008). One of the most common modifications is the N-deacetylation at the C-2. This deletion occurs on MurNAc, GlcNAc or both, mostly in Gram-positive bacteria. GlcNAc deacetylation gives bacteria resistance to lysozyme Amano et al, 1977 and autolysins (bacterial lytic peptidoglycan enzymes). Indeed, lysozyme, and probably autolysins, need to recognize N-acetyl groups in order to cleave the GlcNAc-MurNAc glycosidic bond Vocadlo et al, 2001. Similarly, O-acetylation at C-6 has been observed on MurNAc, and in one case, on GlcNAc Bernard et al., 2011. Bacteria harboring this pattern show an increased virulence Bera et al., 2006, due also to a better resistance to the same peptidoglycan lytic enzyme. Less common or bacterial-species specific glycan modifications can also be found, for which more information is given in Vollmer, 2008 and Moynihan et al, 2014.
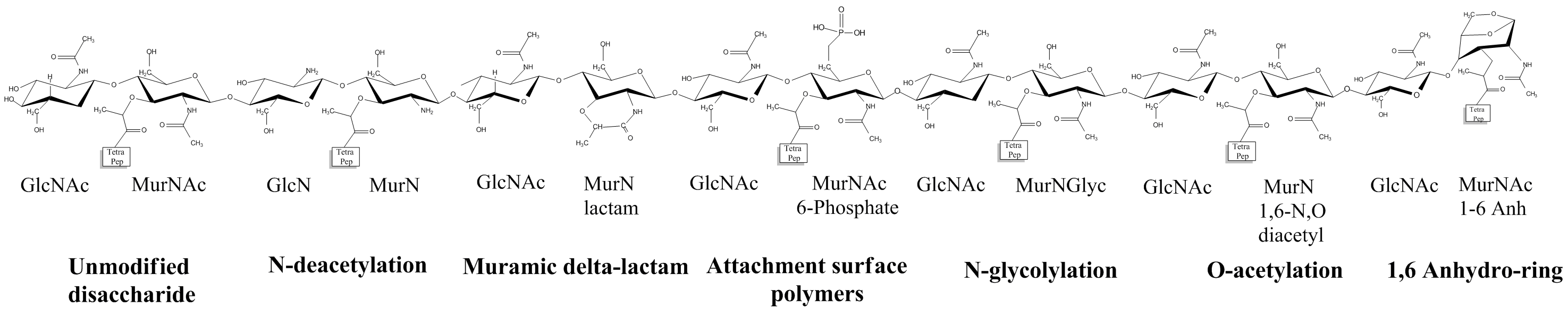
The peptide stems can also be the subject of modifications. Some of them include subtle changes of the α- and ε-carboxylic groups of D-iGlu and m-A2pm through amidation, mostly in Gram-positive bacteria. The α-carboxylic group of D-iGlu can also incorporate additional amino acids and hydroxylation of the carbon chain of the third and fourth residues of the stem peptides can occur at high oxygenation levels (Vollmer, Blanot et al., 2008 for some examples). Others deal with the incorporation of non-canonical D-amino acids (NCDAA), which can be produced by bacteria and released in the extracellular medium. For example, a Vibrio cholerae mutant strain inapt to synthesize peptidoglycan was shown to be able to assimilate in its peptidoglycan D-methionine produced by wild type V. cholerae. In this process, the incorporated atypical residue is exchanged by L,D-transpeptidases with the D-ala in position 4 or 5 of the stem peptide. This mechanism is thought to be important for the cell to adapt to osmotic pressure and to regulate its amount of peptidoglycan Cava et al., 2011.
Peptidoglycan can also be modified by the attachment of macromolecules. As discussed in previous paragraph, diverse glycopolymers are exposed at the surface of Gram-positive bacteria, which requires their binding to the peptidoglycan layer. Most of the time, wall teichoic and teichuronic acids are anchored to the murein by a phosphodiester linkage to the hydroxyl group at the C-6 of MurNAc Vollmer, 2008. In E. coli, Lpp is one of the most abundant peptidoglycan-linked (lipo)protein. While its N-terminal end is anchored to the outer-membrane, its C-terminal extremity can be covalently bound to the peptidoglycan layer to ensure outer-membrane integrity Braun & Rehn, 1969. L,D-transpeptidases were shown to be implied in this reaction, binding the m-A2pm α-carbonyl from a donor peptide stem to the amine group of the side-chain of the Lpp terminal lysine residue Magnet et al., 2007. Due to the L,D-transpeptidases low specificity with respect to different D-amino acids, this process has been recently adapted in vitro to introduce fluorescent D-amino acids (FDAA) in the peptidoglycan and used to label active regions of peptidoglycan synthesis and remodeling in vivo (Kuru et al., 2012Kuru et al., 2015). In Gram-positive bacteria, sortases usually anchor specific proteins to the peptidoglycan and proceed through a transpeptidation reaction between an amino group of the target protein and the peptidoglycan peptide stem. In S. aureus, sortase A was shown to be essential for pathogenesis (for a review on sortases see Bradshaw et al., 2015).
Besides these modifications, peptidoglycan can also been reshaped by a high variety of hydrolases which can virtually cleave almost any bond.
Peptidoglycan composition and all of its modifications are accounting for its unique properties. This enables sacculi to sustain a definite shape for the cell. Most importantly, the rigidity and elasticity of this assembly is essential for bacteria to adapt to the cytoplasmic osmotic pressure and to environmental factors. Due to its role, its unique composition in the living world, and its location outside of the cytoplasmic membrane, peptidoglycan has been and still is an attractive target for antibiotics. However, due to the apparition of resistance to multiple drugs in virtually all bacterial species, it is urgent to develop new strategiesFischbach & Walsh, 2009. This requires a detailed understanding on the biosynthesis and maturation proce>sses involved in the genesis of this essential and ubiquitous biopolymer.
