Bacteria are unicellular organisms, which are spread in a wide variety of environments, from our gut Modi et al., 2014 to the deepest layers of the oceanic crust Mason et al., 2010. These organisms can adopt a wide variety of shapes, from cocci (the spherical Staphylococcus aureus), to bacilli (the rod-like Escherichia coli) or even spirilla (the helically coiled Helicobacter pylori). According to the classification by Carl Woese in 1990 ( Woese et al., 1990), bacteria form one of the two domains, which are constituted by prokaryotes (the second one being archaea). As such, bacteria do not have any nucleus and, with the exception of a few species, do not possess any membrane-delimited organelles, unlike eukaryotes. While the latter have a diameter extended usually from 10 to 100 µm, bacteria are in general one order of magnitude smaller, with a size comprised between 1 and 5 µm (although extreme cases exist, like Thiomargarita namibiensis which can be detected by naked eyes with a maximum diameter of 750 µm).
A schematic representation of the bacterial cell structure is shown in Figure 1, emphasizing on one side the 20- to 530-nm-thick cell envelope (cell wall and capsule) and on another side the cytoplasm, the two parts being delimited by a 70-Å-thick plasma (or cytoplasmic) membrane which consists of a lipid bilayer containing embedded proteins.
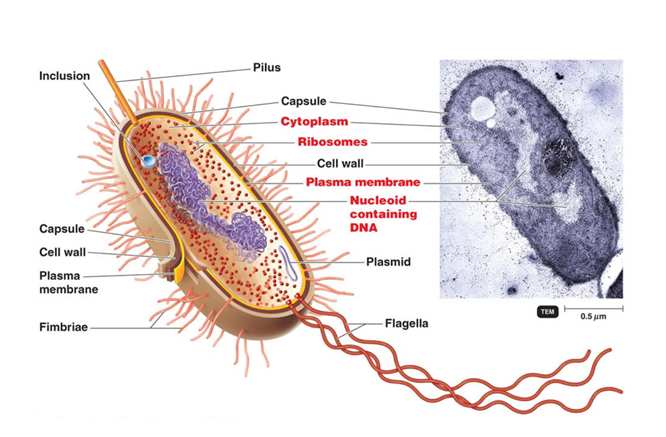
As bacteria do not have membrane-bound organelles, most of the metabolic reactions occur in the cytoplasm. It is also in this space that the genetic information, DeoxyriboNucleic Acid (DNA), is located. Bacterial DNA is usually present as a single, double-stranded, circular chromosome. The cell is relatively small comparatively to its genetic material content (about 4.6 million of base pair for 4400 genes for the bacterial model Escherichia coli), it is therefore compacted by and with its interaction partners in a poorly defined region called the nucleoid. As there are no compartments in the bacterial cell, transcription of DNA in RiboNucleic Acid (RNA) can be coupled to translation into proteins. This last step is performed by 70S ribosomes (denominated from the value of their sedimentation rate by ultracentrifugation, in Svedberg), which are made of two subunits, 30S and 50S. Each of these is composed of ribosomal RNA (rRNA) complexed to numerous proteins. Interestingly, eukaryotic cells have 80S ribosomes with 40S and 60S subunits, which result from different assemblies. These structural divergences enable the specific targeting of the bacterial ribosome with antibiotics without perturbing the patient’s cells. The genome can also be supplemented by one or more smaller circular DNA molecules called plasmids. Contrary to the bacterial chromosome, plasmids are not essential for cell survival but bring additional genes which can be an advantage to cope with environmental stress or to gain resistance (to antibiotics, bacteriophages, and/or chemicals…). This genetic material can be shared rapidly among bacteria by horizontal transfer to promote genetic diversity. This process can occur through three different mechanisms. Some bacteria have the faculty to naturally absorb fragments of DNA, which are present in their environment. This competence is called transformation. They can also directly transfer genes from one to another through a sex pilus, an extended surface structure, by conjugation. Finally, genetic material can be acquired by transduction. In this case, DNA can be conveyed by a bacteriophage from a donor bacterium to a recipient cell.
Although the cytoplasm is lacking of complex internal structures, like a nucleus or a Golgi apparatus, bacteria can sometimes present other elementary compartments in addition to the nucleoid, for which a short and non-exhaustive selection is given here. Among them, thylakoids can be found in cyanobacteria, where they are formed by an ensemble of membranes. Similarly to those observed in chloroplasts, these are the home of photosynthesis and respiration processes. Another compartment involved in converting light into chemical energy is the chlorosome, which is bound to the neighboring cytoplasmic membrane in green sulfur bacteria. It contains large pigment assemblies which are absorbing photons with a very high efficiency to deal with poorly light exposed waters, where these cells are living (see Orf & Blankenship, 2013 for more details). Magnetotactic bacteria are characterized by the presence of magnetosomes, lipid bilayers enclosing each a single magnetic mineral such as magnetite (Fe3O4) or greigite (Fe3S4). The structure obtained is used as a guide for bacteria to follow passively the magnetic field, which may enable these microaerophilic cells (organisms for which oxygen is essential to survive but toxic at atmospheric levels) to find more easily a suitable habitat (for a recent review, see Lower & Bazylinski, 2013). Bacterial organelles can also be only formed by proteins as carboxysomes in cyanobacteria and some other species. These microbodies constitute a shell to likely confine CO2 and its fixing enzyme, Rubisco, in a small volume to favor the latter’s activity Rae et al., 2013.
The bacterial envelope is made of a cell wall layer (detailed in the following section) and various extracellular structures, which are mainly implied in virulence. Among them, one can count the viscous polysaccharidic layer, the capsule that covers the most exposed portion of the bacterial cell. During an infection, this layer enables bacteria to avoid phagocytosis and to resist to antimicrobial peptides and proteins. In addition, the capsule has the faculty to promote adhesion to host cells, other colony individuals, or substrates. This last function can also be fulfilled by excrescences, called pili, and their shorter counterparts fimbriae, which are both resulting from the polymerization of the pilin protein. Flagella are longer hair-like appendages, which are anchored in the cytoplasm and serve as a mean of locomotion. They are the result of a complex assembly of more than 30 proteins using the electrochemical potential difference in protons from both sides of the cytoplasmic membrane to generate motility (reviewed in Morimoto & Minamino, 2014). While these structures are not essential for bacterial survival, the wall delimiting the bacterial cell is fundamental and its chemical structure is highly specific, thus being an attractive target for antibiotics.
