Biomolecular dynamics simulation is a fundamental technology for life sciences research, and its usefulness depends on its accuracy and efficiency. Classical molecular dynamics simulation is fast but lacks chemical accuracy. Quantum chemistry methods such as density functional theory can reach chemical accuracy but cannot scale to support large biomolecules. The authors introduced an artificial intelligence-based ab initio biomolecular dynamics system (AI2BMD) that can efficiently simulate full-atom large biomolecules with ab initio accuracy. AI2BMD uses a protein fragmentation scheme and a machine learning force field to achieve generalizable ab initio accuracy for energy and force calculations for various proteins comprising more than 10,000 atoms. Compared to density functional theory, it reduces the computational time by several orders of magnitude.
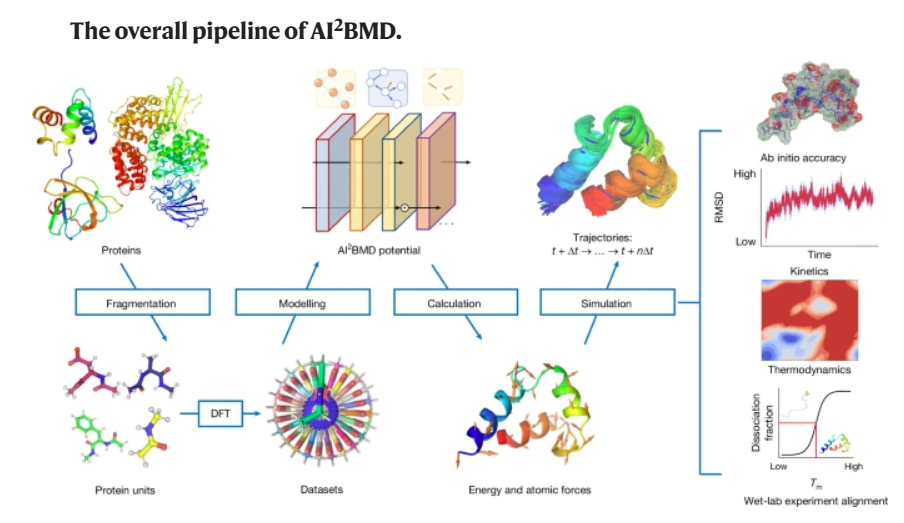
.With several hundred nanoseconds of dynamics simulations, AI2BMD demonstrated its ability to efficiently explore the conformational space of peptides and proteins, deriving accurate 3J couplings that match nuclear magnetic resonance experiments and showing protein folding and unfolding processes. Furthermore, AI2BMD enables precise free-energy calculations for protein folding, and the estimated thermodynamic properties are well aligned with experiments. AI2BMD could potentially complement wet-lab experiments, detect the dynamic processes of bioactivities and enable biomedical research that is currently impossible to conduct.




