Despite ground-breaking innovations in experimental structural biology and protein structure prediction techniques, capturing the structure of the glycans that functionalise proteins remains a challenge. The authors introduce GlycoShape (https://glycoshape.org), an open-access glycan structure database and toolbox designed to restore glycoproteins to their native and functional form in seconds. The GlycoShape database counts over 500 unique glycans, covering the human glycome and augmented by elements from a wide range of organisms, obtained from 1 ms of cumulative sampling from molecular dynamics simulations.
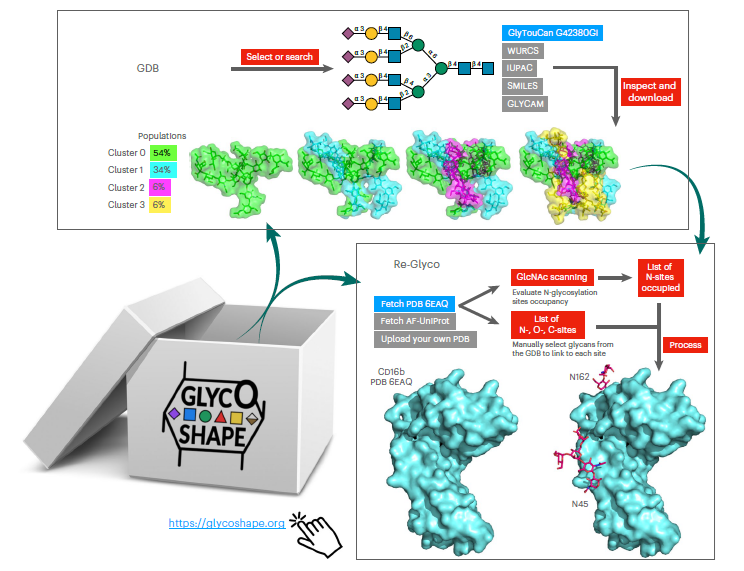
These structures can be linked to proteins with a robust algorithm named Re-Glyco, directly compatible with structural data in open-access repositories, such as the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) and AlphaFold Protein Structure Database, or own. GlycoShape’s quality, performance, and broad applicability are demonstrated by its ability to predict N-glycosylation occupancy. Based on screening all proteins in the PDB with a corresponding glycoproteomics profile, it scored a 93% agreement with the experiment, for a total of 4,259 N–glycosylation sequons




