Advances in molecular dynamics simulations are leading to large increases across spatial, temporal and complexity scale. They provide valuable molecular level insight into processes occurring on the subcellular level. An increasing repertoire of methods to assemble and analyse complex membrane simulations, alongside advances in structural biology methods for membrane proteins, have contributed to our increased understanding of the roles of specific lipid interactions for multiple membrane protein systems.
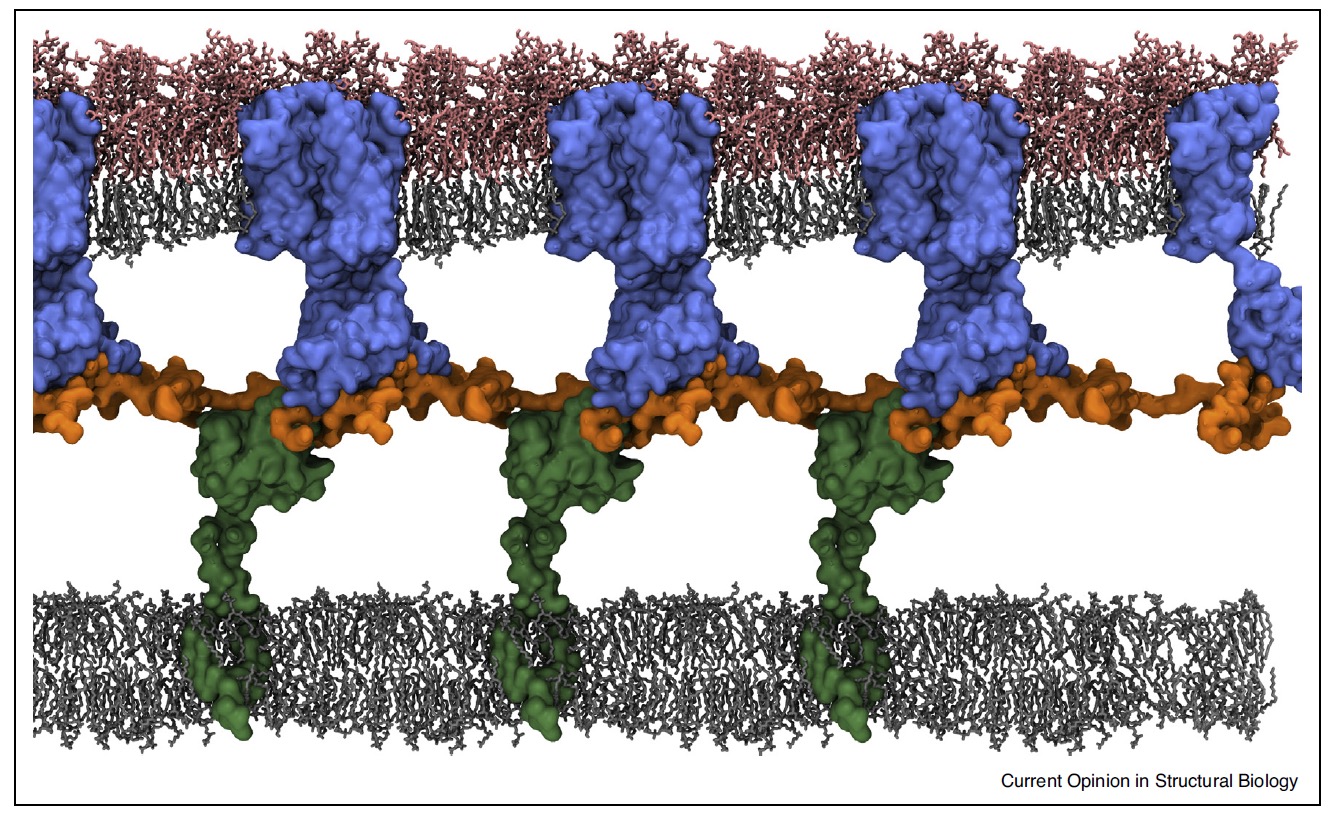
This review is of particular interest for the field of structural glycoscience as well-chosen examples to highlight the structural arrangement of GM1 and GM3 in mammalian membranes and the co-existence of liquid-ordered and liquid-disordered micro-domains. The study confirmed the premise that GMs preferentially partition into the ordered domains. The atomistic simulation of the E.coli cell envelops yields a highly informative view showing the respective locations of lipopolysaccharides, phospholipids, Ompa-full length dime and the TolR.




