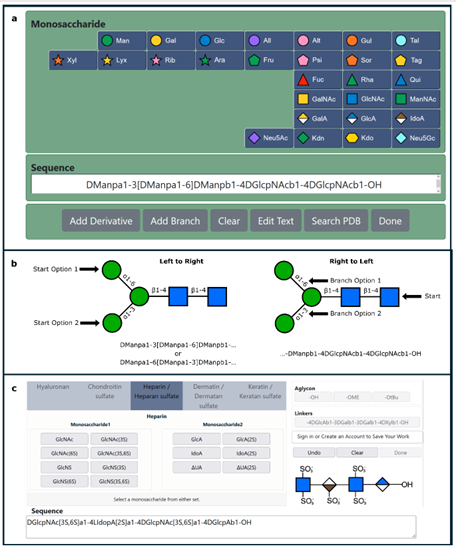
The carbohydrate 3D structure-prediction tools (builders) at GLYCAM-Web (glycam.org) are widely used for generating experimentally-consistent 3D structures of oligosaccharides suitable for data interpretation, hypothesis generation, simple visualization, and subsequent molecular dynamics (MD) simulation. The graphical user interface (GUI) enables users to create carbohydrate sequences (e.g. DGalpb1-4DGlcpb1-OH) that are converted to 3D models of the carbohydrate structures in multiple formats, including PDB and OFF (AMBER software format). The resulting structures are energy minimized before download and online visualization. There are advanced options for selecting which shapes (rotamers) of the oligosaccharide to generate, and for creating explicitly solvated structures for subsequent MD simulation. The GLYCAM-Web builders integrate known conformational preferences of oligosaccharides and employ the GLYCAM forcefield for energy minimization with algorithms tailored for speed and scalability. Even for large oligosaccharides (100 residues, ~2100 atoms), a 3D structure is typically returned to the user in less than a minute.