Protein glycosylation is a general post-translational modification involved in cell membrane formation, is crucial to dictate proper conformation of many membrane proteins, retaining stability on some secreted glycoproteins, and playing a role in cell–cell adhesion. Therefore, the analysis of post-translational modifications of proteins, including glycosylation, is essential to understand the exact functions and interactions of each protein molecule. To conduct large-scale analyses more efficiently, it is essential to promote the accumulation, sharing, and reuse of experimental and analytical data in accordance with the FAIR (Findability, Accessibility, Interoperability, and Re-usability) data principles. However, there was no FAIR data repository for storing and sharing information on glycoconjugates, including glycopeptides and glycoproteins, in a standardised format.
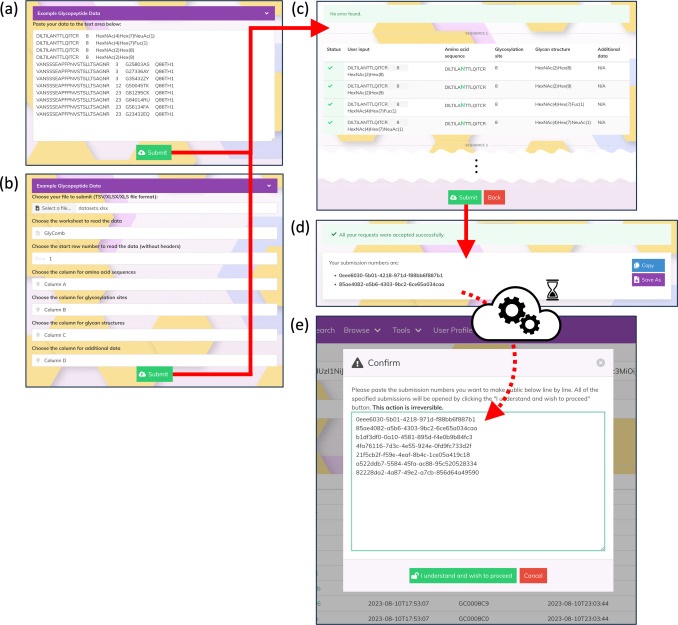
The authors have developed GlyComb (https://glycomb.glycosmos.org) as a new standardized data repository for glycoconjugate data. GlyComb can assign a unique identifier to a set of glycosylation information associated with a specific peptide sequence or UniProt ID. By standardising glycosylation data using GlyComb identifiers and coordinating with existing web resources such as GlyTouCan and GlycoPOST, a comprehensive system for data submission and sharing between researchers can be established. This article describes how GlyComb can integrate the variety of glycoconjugate data already registered in existing data repositories to gain a better understanding of the available glycopeptides and glycoproteins and their glycosylation patterns. We also explain how this system can be used as a basis for a better understanding of glycan function.