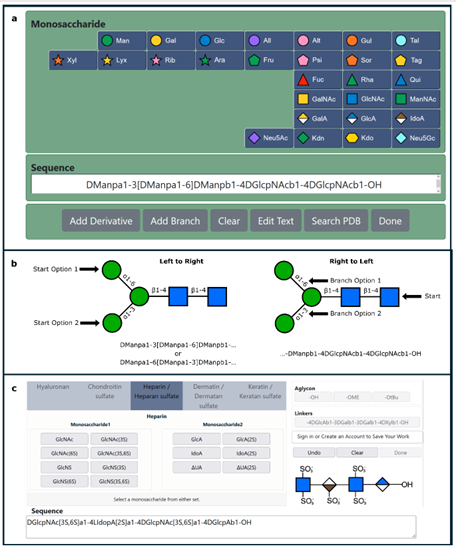
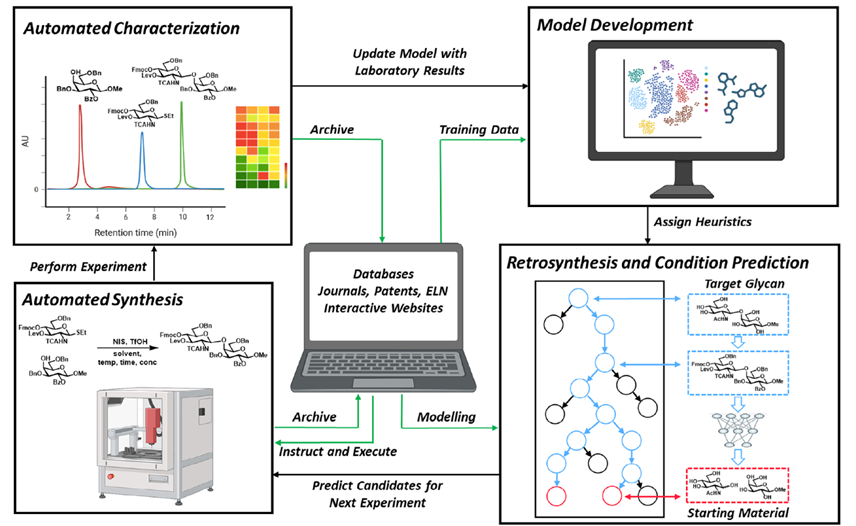
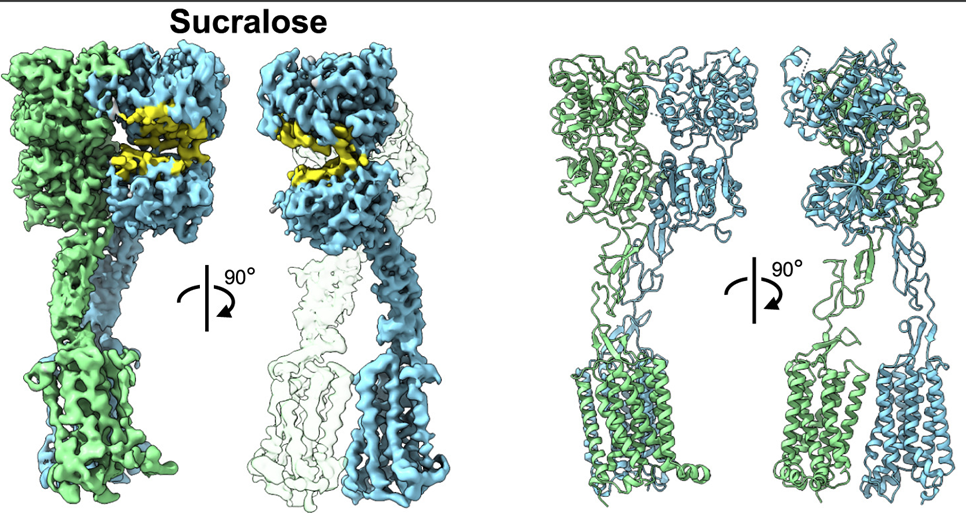
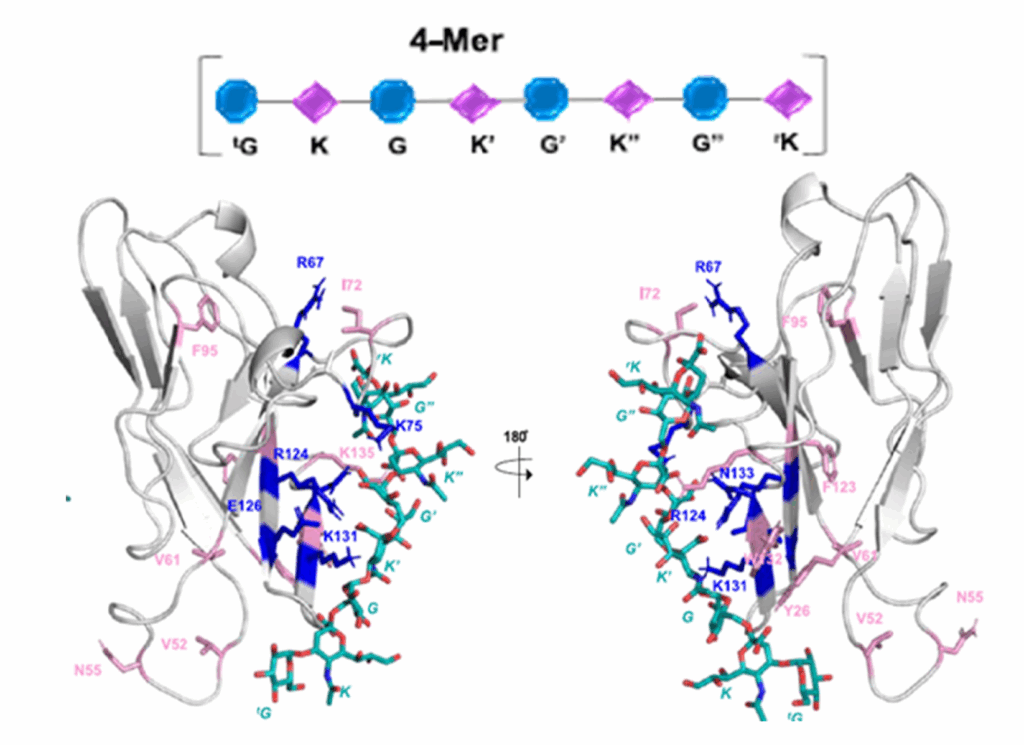
DeepMind’s program, called AlphaFold, outperformed approximately 100 other teams in a biennial protein-structure prediction challenge called CASP, short for Critical Assessment of Structure Prediction. DeepMind’s AlphaFold program has determined the shape of round two-thirds of the proteins with accuracy comparable to laboratory experiments. AlphaFold’s accuracy with most of the other proteins was also high, though not quite at that level.” predictioncenter.org/casp14 /doc/CASP14_press_release.html. Such a gigantic leap offers the community of molecular biologist new horizons to contemplate and tackle. The ability to compute a protein’s shape from its genetic code would accelerate research in many areas. It is also appropriate to look back and analyse the key steps and how the folding problem has concentrated so much involvement and development during 60 years. (J.R. Helliwell, IUCr Newslletter (2020) 28 https://www.iucr.org/news/newsletter/volume-28/number-4/deepmind-an…) ; It must be recognized that AlphaFold success partially relies upon almost 200,000 protein structures that have been experimentally determined, and publicly available. The major strength of DeepMind is the breath of other areas of research involving Artificial Intelligence.
8-minute video from DeepMind on their work is to be found