Lectins are non-covalent glycan-binding proteins mediating cellular interactions but their annotation in newly sequenced organisms is lacking. The limited size of functional domains and the low level of sequence similarity challenge usual bioinformatics tools. The identification of lectin domains in proteomes requires the manual curation of sequence alignments based on structural folds.
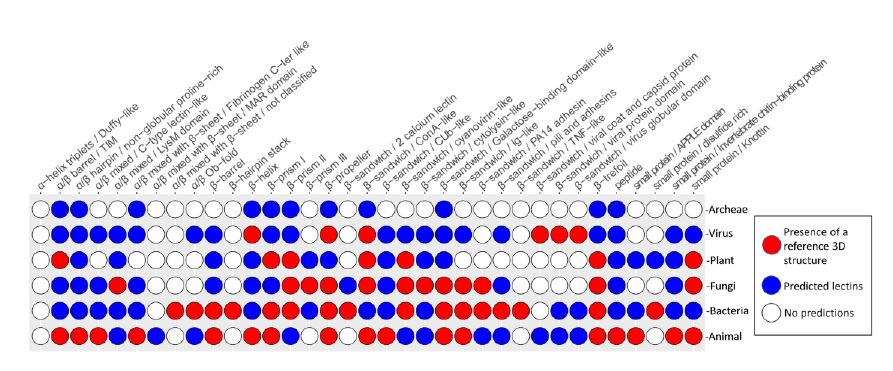
A new lectin classification is proposed. It is built on three levels: 1) 35 lectin domain folds, 2) 109 classes of lectins sharing at least 20% sequence similarity and 3) 350 families of lectins sharing at least 70% sequence similarity. This information is compiled in the UniLectin platform that includes the previously described UniLectin3D database of curated lectin 3D structures. Since its first release, UniLectin3D has been updated with 485 additional 3D structures. The database is now complemented by two additional modules: PropLec containing predicted β-propeller lectins and LectomeXplore including predicted lectins for every curated lectin class. UniLectin is accessible at https://www.unilectin.eu/