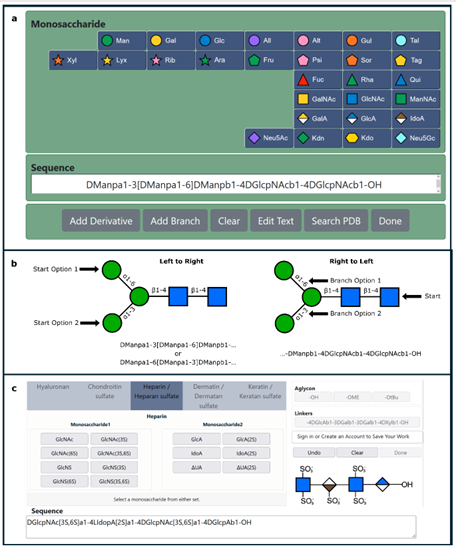
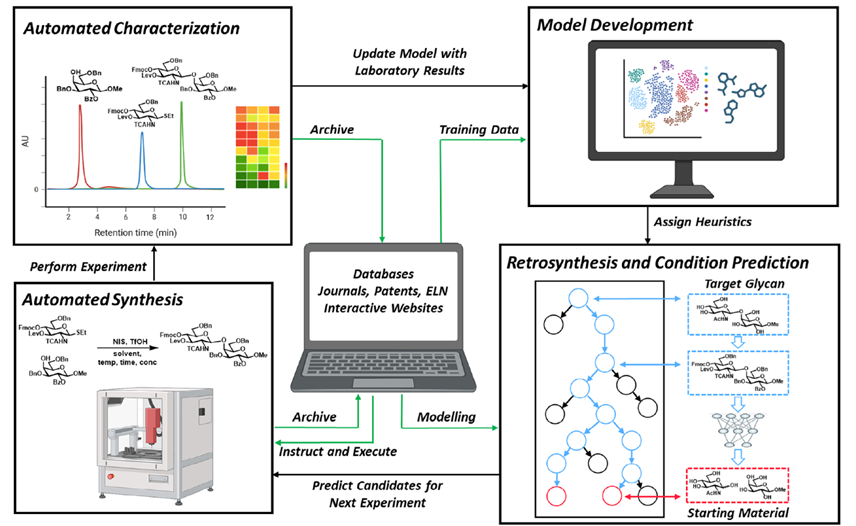
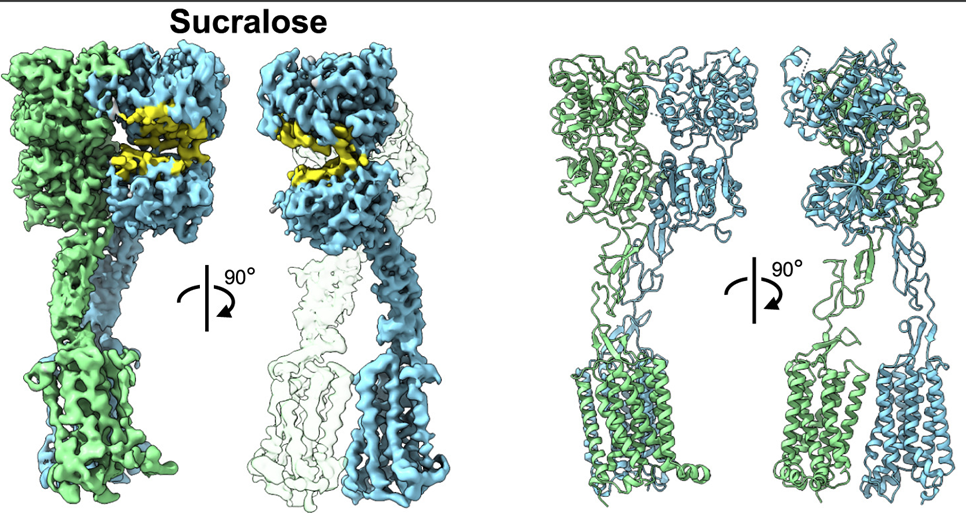
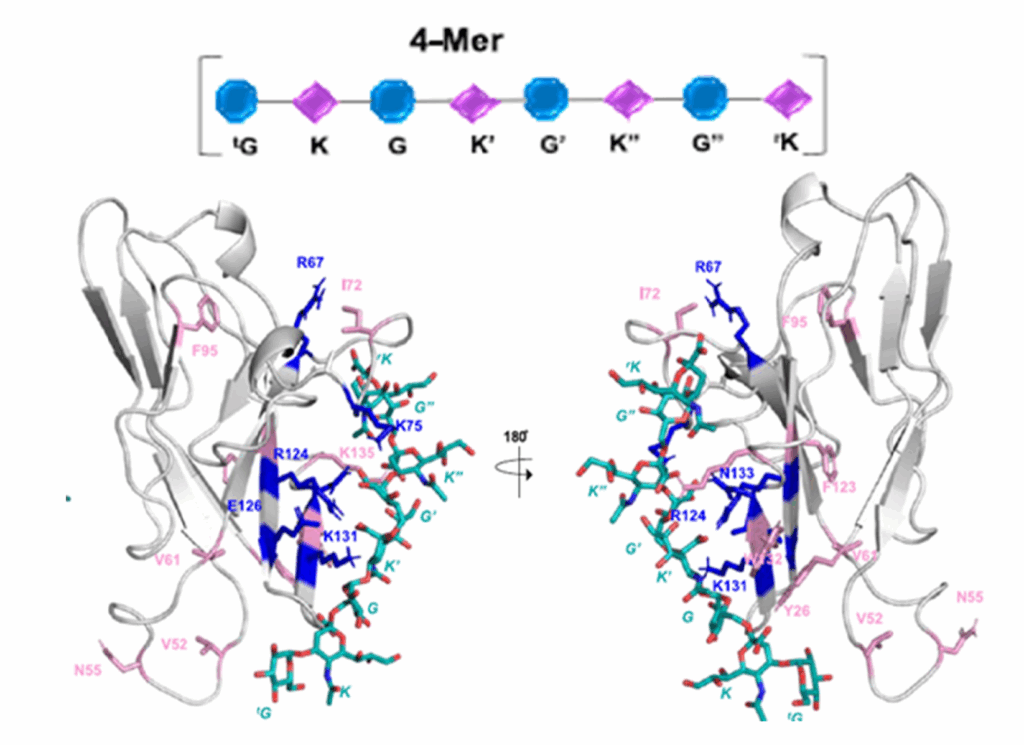
Protein structure prediction can be used to determine the three-dimensional shape of a protein from its amino acid sequence1. This problem is of fundamental importance as the structure of a protein largely determines its function; however, protein structures can be difficult to determine experimentally. Considerable progress has recently been made by leveraging genetic information.
It is possible to infer which amino acid residues are in contact by analysing covariation in homologous sequences, which aids in the prediction of protein structures. The idea is predicated on the following premise: if two amino-acid residues in a protein are close together in 3D space, then a mutation that replaces one of them with a different residue (for example, large for small) will probably induce, at a later time, a mutation that alters the other residue in a compensatory direction (in our example, swapping small for large). The set of co-evolving residues, therefore, encodes valuable spatial information and can be found by analysing the sequences of evolutionarily related proteins.
AlphaFold predicts the probabilities of residues being separated by different distances. Because probabilities and energies are interconvertible, AlphaFold predicts an energy landscape — one that overlaps in its lowest basin with the true landscape but is much smoother. In fact, AlphaFold’s landscape is so smooth that it nearly eliminates the need for searching. This makes it possible to use a simple procedure to find the most favourable conformation, rather than the complex search algorithms employed by other methods.
Neural network can be trained to make accurate predictions of the distances between pairs of residues, which convey more information about the structure than contact predictions. Using this information, a potential of mean force can be constructed that accurately describe the shape of a protein. We find that the resulting potential can be optimized by a simple gradient descent algorithm to generate structures without complex sampling procedures.
The resulting algorithm outperformed all entrants at the most recent blind assessment of methods used to predict protein structures (the CASP13 event), generating the best structure for 25 out of 43 proteins, compared with 3 out of 43 for the next-best method. AlphaFold’s predictions had a median accuracy of 6.6 ångströms. AlphaFold represents a considerable advance in protein-structure prediction.
AlphaFold is not yet accurate enough for most applications, such as working out the catalytic mechanisms of enzymes or how drugs bind to proteins (which both typically require 2–3 Å resolution). And although AlphaFold’s search procedure is much simpler than most modern methods, it can still be slow, taking tens to hundreds of hours to make a single prediction.